
Hubei virga-like virus 15
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
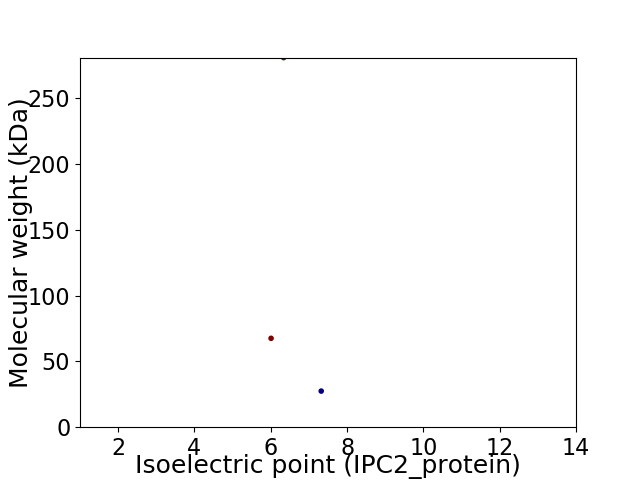
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KJT4|A0A1L3KJT4_9VIRU Non-structural protein 2 OS=Hubei virga-like virus 15 OX=1923330 PE=4 SV=1
MM1 pKa = 7.48LVFNFLLLFTSTVAVHH17 pKa = 6.7RR18 pKa = 11.84PIPNYY23 pKa = 10.92DD24 pKa = 3.62PDD26 pKa = 3.99DD27 pKa = 4.62KK28 pKa = 11.27FWFSPPSADD37 pKa = 3.93AGKK40 pKa = 8.45LTKK43 pKa = 9.86MYY45 pKa = 10.21DD46 pKa = 3.15LHH48 pKa = 8.67GGFKK52 pKa = 8.86WCHH55 pKa = 5.1GSVLRR60 pKa = 11.84YY61 pKa = 9.62GFWEE65 pKa = 4.37GLNAFTIYY73 pKa = 9.83MYY75 pKa = 10.81LEE77 pKa = 4.54KK78 pKa = 10.72DD79 pKa = 3.68LYY81 pKa = 10.67RR82 pKa = 11.84LRR84 pKa = 11.84NKK86 pKa = 8.43VTIHH90 pKa = 6.17TGQYY94 pKa = 9.5VYY96 pKa = 11.33SLVGAHH102 pKa = 7.31LLTNDD107 pKa = 4.27RR108 pKa = 11.84EE109 pKa = 4.39FEE111 pKa = 4.22NFNSKK116 pKa = 10.4HH117 pKa = 6.04SLACFPDD124 pKa = 3.39TGLPKK129 pKa = 9.77CTLRR133 pKa = 11.84EE134 pKa = 3.66ICKK137 pKa = 10.32ARR139 pKa = 11.84TGKK142 pKa = 10.56KK143 pKa = 9.58PFPLVGTCEE152 pKa = 4.09NKK154 pKa = 10.25DD155 pKa = 3.35FRR157 pKa = 11.84IVEE160 pKa = 4.29RR161 pKa = 11.84GADD164 pKa = 3.77FDD166 pKa = 4.46PQCVCYY172 pKa = 10.45GDD174 pKa = 4.95CDD176 pKa = 3.94HH177 pKa = 7.46DD178 pKa = 4.57CEE180 pKa = 5.51HH181 pKa = 6.47STLHH185 pKa = 5.95NYY187 pKa = 9.99KK188 pKa = 10.56GIEE191 pKa = 3.86FSLGYY196 pKa = 9.92RR197 pKa = 11.84FNSKK201 pKa = 10.35PPLKK205 pKa = 10.74VSVDD209 pKa = 3.65RR210 pKa = 11.84YY211 pKa = 10.88GVGCIDD217 pKa = 4.0GLTSLAHH224 pKa = 6.38TYY226 pKa = 8.1WQTANYY232 pKa = 9.2HH233 pKa = 5.79PAYY236 pKa = 9.7LANSGVLQTFRR247 pKa = 11.84LRR249 pKa = 11.84KK250 pKa = 9.24EE251 pKa = 4.2FEE253 pKa = 4.15DD254 pKa = 4.65LRR256 pKa = 11.84PTVLNVTISNRR267 pKa = 11.84YY268 pKa = 7.92VCDD271 pKa = 3.45DD272 pKa = 3.24AMSRR276 pKa = 11.84RR277 pKa = 11.84YY278 pKa = 9.07HH279 pKa = 6.55ALDD282 pKa = 3.5GVVIYY287 pKa = 7.3PTAIDD292 pKa = 3.6CTEE295 pKa = 4.04FVPFDD300 pKa = 3.62FHH302 pKa = 7.27TLICKK307 pKa = 10.0DD308 pKa = 3.28ALFVPMCPTEE318 pKa = 3.99VFKK321 pKa = 11.26PMVARR326 pKa = 11.84YY327 pKa = 9.19RR328 pKa = 11.84YY329 pKa = 10.24YY330 pKa = 10.78DD331 pKa = 3.26ITNHH335 pKa = 5.43RR336 pKa = 11.84RR337 pKa = 11.84VVGALDD343 pKa = 3.7LVGAIQNQTGDD354 pKa = 3.37TMVDD358 pKa = 2.95HH359 pKa = 7.0FFDD362 pKa = 3.98HH363 pKa = 6.86FAGNATRR370 pKa = 11.84HH371 pKa = 4.9HH372 pKa = 5.53VVRR375 pKa = 11.84RR376 pKa = 11.84SISDD380 pKa = 3.86WISGVVEE387 pKa = 4.55KK388 pKa = 10.89LFEE391 pKa = 4.93PIKK394 pKa = 10.81YY395 pKa = 9.61LVQEE399 pKa = 4.15ILDD402 pKa = 4.74LIRR405 pKa = 11.84PILVEE410 pKa = 3.55IAGEE414 pKa = 4.06IFKK417 pKa = 10.98LILDD421 pKa = 3.73ILFDD425 pKa = 4.13LVGILDD431 pKa = 3.98SLLKK435 pKa = 10.68KK436 pKa = 10.91LEE438 pKa = 4.16DD439 pKa = 3.76QIHH442 pKa = 6.08NLVDD446 pKa = 5.1LIRR449 pKa = 11.84VLLHH453 pKa = 6.27HH454 pKa = 7.13LFSIVTRR461 pKa = 11.84LLLSLEE467 pKa = 4.21CSYY470 pKa = 11.59HH471 pKa = 7.51LFEE474 pKa = 6.38ALFLVAFFRR483 pKa = 11.84YY484 pKa = 8.67IFHH487 pKa = 7.24SIYY490 pKa = 10.11IASLLTFFIFLIIGFEE506 pKa = 4.25RR507 pKa = 11.84IYY509 pKa = 10.11PSPVYY514 pKa = 10.48YY515 pKa = 10.58YY516 pKa = 10.74LSEE519 pKa = 4.12KK520 pKa = 10.3FRR522 pKa = 11.84LSVCSVPVRR531 pKa = 11.84YY532 pKa = 10.12LMNLQGAYY540 pKa = 10.05AVLKK544 pKa = 10.78DD545 pKa = 3.59LFEE548 pKa = 4.36YY549 pKa = 10.95LSGFMPNFLYY559 pKa = 10.48QGSYY563 pKa = 11.28DD564 pKa = 3.83LFIIASLMVSAFIISFFFSDD584 pKa = 3.34VV585 pKa = 3.09
MM1 pKa = 7.48LVFNFLLLFTSTVAVHH17 pKa = 6.7RR18 pKa = 11.84PIPNYY23 pKa = 10.92DD24 pKa = 3.62PDD26 pKa = 3.99DD27 pKa = 4.62KK28 pKa = 11.27FWFSPPSADD37 pKa = 3.93AGKK40 pKa = 8.45LTKK43 pKa = 9.86MYY45 pKa = 10.21DD46 pKa = 3.15LHH48 pKa = 8.67GGFKK52 pKa = 8.86WCHH55 pKa = 5.1GSVLRR60 pKa = 11.84YY61 pKa = 9.62GFWEE65 pKa = 4.37GLNAFTIYY73 pKa = 9.83MYY75 pKa = 10.81LEE77 pKa = 4.54KK78 pKa = 10.72DD79 pKa = 3.68LYY81 pKa = 10.67RR82 pKa = 11.84LRR84 pKa = 11.84NKK86 pKa = 8.43VTIHH90 pKa = 6.17TGQYY94 pKa = 9.5VYY96 pKa = 11.33SLVGAHH102 pKa = 7.31LLTNDD107 pKa = 4.27RR108 pKa = 11.84EE109 pKa = 4.39FEE111 pKa = 4.22NFNSKK116 pKa = 10.4HH117 pKa = 6.04SLACFPDD124 pKa = 3.39TGLPKK129 pKa = 9.77CTLRR133 pKa = 11.84EE134 pKa = 3.66ICKK137 pKa = 10.32ARR139 pKa = 11.84TGKK142 pKa = 10.56KK143 pKa = 9.58PFPLVGTCEE152 pKa = 4.09NKK154 pKa = 10.25DD155 pKa = 3.35FRR157 pKa = 11.84IVEE160 pKa = 4.29RR161 pKa = 11.84GADD164 pKa = 3.77FDD166 pKa = 4.46PQCVCYY172 pKa = 10.45GDD174 pKa = 4.95CDD176 pKa = 3.94HH177 pKa = 7.46DD178 pKa = 4.57CEE180 pKa = 5.51HH181 pKa = 6.47STLHH185 pKa = 5.95NYY187 pKa = 9.99KK188 pKa = 10.56GIEE191 pKa = 3.86FSLGYY196 pKa = 9.92RR197 pKa = 11.84FNSKK201 pKa = 10.35PPLKK205 pKa = 10.74VSVDD209 pKa = 3.65RR210 pKa = 11.84YY211 pKa = 10.88GVGCIDD217 pKa = 4.0GLTSLAHH224 pKa = 6.38TYY226 pKa = 8.1WQTANYY232 pKa = 9.2HH233 pKa = 5.79PAYY236 pKa = 9.7LANSGVLQTFRR247 pKa = 11.84LRR249 pKa = 11.84KK250 pKa = 9.24EE251 pKa = 4.2FEE253 pKa = 4.15DD254 pKa = 4.65LRR256 pKa = 11.84PTVLNVTISNRR267 pKa = 11.84YY268 pKa = 7.92VCDD271 pKa = 3.45DD272 pKa = 3.24AMSRR276 pKa = 11.84RR277 pKa = 11.84YY278 pKa = 9.07HH279 pKa = 6.55ALDD282 pKa = 3.5GVVIYY287 pKa = 7.3PTAIDD292 pKa = 3.6CTEE295 pKa = 4.04FVPFDD300 pKa = 3.62FHH302 pKa = 7.27TLICKK307 pKa = 10.0DD308 pKa = 3.28ALFVPMCPTEE318 pKa = 3.99VFKK321 pKa = 11.26PMVARR326 pKa = 11.84YY327 pKa = 9.19RR328 pKa = 11.84YY329 pKa = 10.24YY330 pKa = 10.78DD331 pKa = 3.26ITNHH335 pKa = 5.43RR336 pKa = 11.84RR337 pKa = 11.84VVGALDD343 pKa = 3.7LVGAIQNQTGDD354 pKa = 3.37TMVDD358 pKa = 2.95HH359 pKa = 7.0FFDD362 pKa = 3.98HH363 pKa = 6.86FAGNATRR370 pKa = 11.84HH371 pKa = 4.9HH372 pKa = 5.53VVRR375 pKa = 11.84RR376 pKa = 11.84SISDD380 pKa = 3.86WISGVVEE387 pKa = 4.55KK388 pKa = 10.89LFEE391 pKa = 4.93PIKK394 pKa = 10.81YY395 pKa = 9.61LVQEE399 pKa = 4.15ILDD402 pKa = 4.74LIRR405 pKa = 11.84PILVEE410 pKa = 3.55IAGEE414 pKa = 4.06IFKK417 pKa = 10.98LILDD421 pKa = 3.73ILFDD425 pKa = 4.13LVGILDD431 pKa = 3.98SLLKK435 pKa = 10.68KK436 pKa = 10.91LEE438 pKa = 4.16DD439 pKa = 3.76QIHH442 pKa = 6.08NLVDD446 pKa = 5.1LIRR449 pKa = 11.84VLLHH453 pKa = 6.27HH454 pKa = 7.13LFSIVTRR461 pKa = 11.84LLLSLEE467 pKa = 4.21CSYY470 pKa = 11.59HH471 pKa = 7.51LFEE474 pKa = 6.38ALFLVAFFRR483 pKa = 11.84YY484 pKa = 8.67IFHH487 pKa = 7.24SIYY490 pKa = 10.11IASLLTFFIFLIIGFEE506 pKa = 4.25RR507 pKa = 11.84IYY509 pKa = 10.11PSPVYY514 pKa = 10.48YY515 pKa = 10.58YY516 pKa = 10.74LSEE519 pKa = 4.12KK520 pKa = 10.3FRR522 pKa = 11.84LSVCSVPVRR531 pKa = 11.84YY532 pKa = 10.12LMNLQGAYY540 pKa = 10.05AVLKK544 pKa = 10.78DD545 pKa = 3.59LFEE548 pKa = 4.36YY549 pKa = 10.95LSGFMPNFLYY559 pKa = 10.48QGSYY563 pKa = 11.28DD564 pKa = 3.83LFIIASLMVSAFIISFFFSDD584 pKa = 3.34VV585 pKa = 3.09
Molecular weight: 67.6 kDa
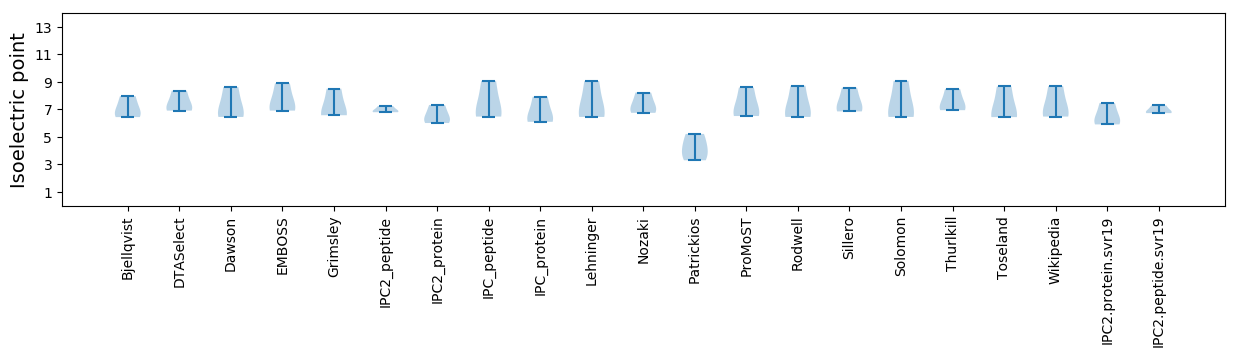
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KK08|A0A1L3KK08_9VIRU Uncharacterized protein OS=Hubei virga-like virus 15 OX=1923330 PE=4 SV=1
MM1 pKa = 7.53SSADD5 pKa = 3.73RR6 pKa = 11.84DD7 pKa = 3.83DD8 pKa = 3.8ARR10 pKa = 11.84TRR12 pKa = 11.84MVDD15 pKa = 2.83SRR17 pKa = 11.84GITTIKK23 pKa = 10.45GGNLTGATLIQSVQDD38 pKa = 3.27AYY40 pKa = 10.29TNVMQNAIATSMMALLVIITLAEE63 pKa = 3.97HH64 pKa = 6.57AQTKK68 pKa = 10.44GPLEE72 pKa = 4.27SMRR75 pKa = 11.84DD76 pKa = 3.4ALAALVDD83 pKa = 3.91SGEE86 pKa = 4.02MSGAKK91 pKa = 10.09NFIASAVHH99 pKa = 6.4NILVYY104 pKa = 10.2VVNHH108 pKa = 6.77KK109 pKa = 10.77DD110 pKa = 3.63VIIEE114 pKa = 4.26VAWIWWPWVVKK125 pKa = 10.34PSTKK129 pKa = 10.5NMWMSVVLSFFAVFAEE145 pKa = 4.14LTLFEE150 pKa = 4.65IVALANAYY158 pKa = 8.01FLWVSLRR165 pKa = 11.84NPAHH169 pKa = 6.31KK170 pKa = 9.07TLAFIFGMVILVVSLEE186 pKa = 4.03FNLTKK191 pKa = 10.36EE192 pKa = 4.31KK193 pKa = 10.53PLTPTDD199 pKa = 3.44IPHH202 pKa = 6.56TFDD205 pKa = 4.25PFPNVKK211 pKa = 10.36NPDD214 pKa = 3.99DD215 pKa = 4.72RR216 pKa = 11.84PPAPDD221 pKa = 3.35FATITKK227 pKa = 10.26KK228 pKa = 10.49EE229 pKa = 4.01GARR232 pKa = 11.84VARR235 pKa = 11.84SIGSGTIPPPTKK247 pKa = 9.99PFKK250 pKa = 10.87
MM1 pKa = 7.53SSADD5 pKa = 3.73RR6 pKa = 11.84DD7 pKa = 3.83DD8 pKa = 3.8ARR10 pKa = 11.84TRR12 pKa = 11.84MVDD15 pKa = 2.83SRR17 pKa = 11.84GITTIKK23 pKa = 10.45GGNLTGATLIQSVQDD38 pKa = 3.27AYY40 pKa = 10.29TNVMQNAIATSMMALLVIITLAEE63 pKa = 3.97HH64 pKa = 6.57AQTKK68 pKa = 10.44GPLEE72 pKa = 4.27SMRR75 pKa = 11.84DD76 pKa = 3.4ALAALVDD83 pKa = 3.91SGEE86 pKa = 4.02MSGAKK91 pKa = 10.09NFIASAVHH99 pKa = 6.4NILVYY104 pKa = 10.2VVNHH108 pKa = 6.77KK109 pKa = 10.77DD110 pKa = 3.63VIIEE114 pKa = 4.26VAWIWWPWVVKK125 pKa = 10.34PSTKK129 pKa = 10.5NMWMSVVLSFFAVFAEE145 pKa = 4.14LTLFEE150 pKa = 4.65IVALANAYY158 pKa = 8.01FLWVSLRR165 pKa = 11.84NPAHH169 pKa = 6.31KK170 pKa = 9.07TLAFIFGMVILVVSLEE186 pKa = 4.03FNLTKK191 pKa = 10.36EE192 pKa = 4.31KK193 pKa = 10.53PLTPTDD199 pKa = 3.44IPHH202 pKa = 6.56TFDD205 pKa = 4.25PFPNVKK211 pKa = 10.36NPDD214 pKa = 3.99DD215 pKa = 4.72RR216 pKa = 11.84PPAPDD221 pKa = 3.35FATITKK227 pKa = 10.26KK228 pKa = 10.49EE229 pKa = 4.01GARR232 pKa = 11.84VARR235 pKa = 11.84SIGSGTIPPPTKK247 pKa = 9.99PFKK250 pKa = 10.87
Molecular weight: 27.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3289 |
250 |
2454 |
1096.3 |
125.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.142 ± 0.855 | 2.128 ± 0.391 |
6.811 ± 0.377 | 5.594 ± 0.727 |
5.655 ± 0.751 | 5.838 ± 0.39 |
2.402 ± 0.443 | 6.233 ± 0.241 |
5.564 ± 0.394 | 9.03 ± 0.889 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.01 ± 0.365 | 4.165 ± 0.211 |
3.922 ± 0.635 | 2.067 ± 0.173 |
6.476 ± 0.804 | 5.929 ± 0.087 |
5.442 ± 0.47 | 7.662 ± 0.315 |
0.973 ± 0.275 | 4.926 ± 0.685 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
