
Nitrosopumilaceae archaeon
Taxonomy: cellular organisms; Archaea; TACK group; Thaumarchaeota; Nitrosopumilales; Nitrosopumilaceae; unclassified Nitrosopumilaceae
Average proteome isoelectric point is 6.66
Get precalculated fractions of proteins
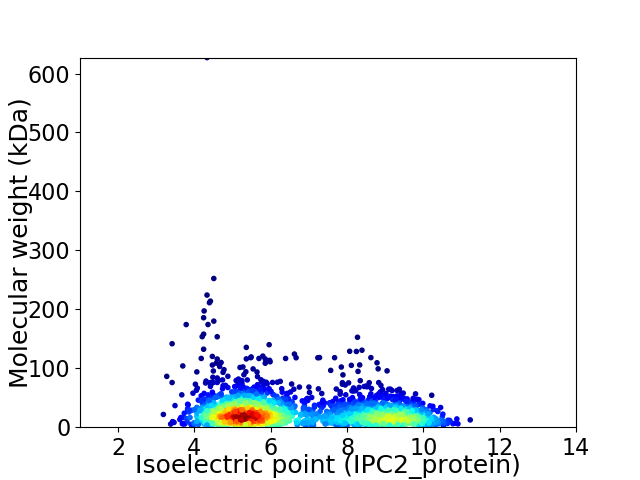
Virtual 2D-PAGE plot for 2237 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A447G2S2|A0A447G2S2_9ARCH Uncharacterized protein OS=Nitrosopumilaceae archaeon OX=2202732 GN=IBTHAUMO2_190018 PE=4 SV=1
MM1 pKa = 6.76KK2 pKa = 9.23TAIIAGIVAAAAAVGVAAVLAGMGPDD28 pKa = 3.42QMDD31 pKa = 3.84GQMMADD37 pKa = 3.65EE38 pKa = 4.49MADD41 pKa = 3.28GGMMADD47 pKa = 3.98EE48 pKa = 4.35MAGGITVAASFLPIYY63 pKa = 10.13EE64 pKa = 4.09FATAVGGNTVSVSQLVPPGAEE85 pKa = 3.74PHH87 pKa = 6.59DD88 pKa = 3.89WEE90 pKa = 4.23PTVRR94 pKa = 11.84QVQEE98 pKa = 3.96LQEE101 pKa = 4.53ADD103 pKa = 3.18IIAINGIHH111 pKa = 5.91FEE113 pKa = 4.1EE114 pKa = 4.55WLGDD118 pKa = 3.37LDD120 pKa = 4.56AYY122 pKa = 10.65GGALVDD128 pKa = 3.63TSAPVEE134 pKa = 4.34VMHH137 pKa = 6.98EE138 pKa = 4.42DD139 pKa = 3.2PHH141 pKa = 7.71IWLVPTNAALQARR154 pKa = 11.84EE155 pKa = 3.66IAMALAEE162 pKa = 4.67IDD164 pKa = 3.73PGNADD169 pKa = 3.72EE170 pKa = 4.71YY171 pKa = 10.72MSNADD176 pKa = 3.13IFTAEE181 pKa = 4.21MDD183 pKa = 4.41ALDD186 pKa = 3.94ADD188 pKa = 3.53IRR190 pKa = 11.84AGLEE194 pKa = 3.93GCGTEE199 pKa = 4.57FVTAHH204 pKa = 6.26GAFSYY209 pKa = 10.31FAEE212 pKa = 4.7EE213 pKa = 4.23YY214 pKa = 11.07GLVQHH219 pKa = 7.12AIIAGGDD226 pKa = 3.49PHH228 pKa = 8.74GEE230 pKa = 3.95ASPHH234 pKa = 7.08DD235 pKa = 3.97IGDD238 pKa = 4.14AAAHH242 pKa = 5.84AAEE245 pKa = 4.32TGYY248 pKa = 10.51ILAEE252 pKa = 4.25SPSRR256 pKa = 11.84LVDD259 pKa = 3.38ATAAEE264 pKa = 4.4AGAEE268 pKa = 4.04VLILDD273 pKa = 4.38PLEE276 pKa = 4.41TSEE279 pKa = 5.41GPYY282 pKa = 9.78EE283 pKa = 4.07DD284 pKa = 3.29VMRR287 pKa = 11.84RR288 pKa = 11.84NLGVLQQALCTQQ300 pKa = 3.87
MM1 pKa = 6.76KK2 pKa = 9.23TAIIAGIVAAAAAVGVAAVLAGMGPDD28 pKa = 3.42QMDD31 pKa = 3.84GQMMADD37 pKa = 3.65EE38 pKa = 4.49MADD41 pKa = 3.28GGMMADD47 pKa = 3.98EE48 pKa = 4.35MAGGITVAASFLPIYY63 pKa = 10.13EE64 pKa = 4.09FATAVGGNTVSVSQLVPPGAEE85 pKa = 3.74PHH87 pKa = 6.59DD88 pKa = 3.89WEE90 pKa = 4.23PTVRR94 pKa = 11.84QVQEE98 pKa = 3.96LQEE101 pKa = 4.53ADD103 pKa = 3.18IIAINGIHH111 pKa = 5.91FEE113 pKa = 4.1EE114 pKa = 4.55WLGDD118 pKa = 3.37LDD120 pKa = 4.56AYY122 pKa = 10.65GGALVDD128 pKa = 3.63TSAPVEE134 pKa = 4.34VMHH137 pKa = 6.98EE138 pKa = 4.42DD139 pKa = 3.2PHH141 pKa = 7.71IWLVPTNAALQARR154 pKa = 11.84EE155 pKa = 3.66IAMALAEE162 pKa = 4.67IDD164 pKa = 3.73PGNADD169 pKa = 3.72EE170 pKa = 4.71YY171 pKa = 10.72MSNADD176 pKa = 3.13IFTAEE181 pKa = 4.21MDD183 pKa = 4.41ALDD186 pKa = 3.94ADD188 pKa = 3.53IRR190 pKa = 11.84AGLEE194 pKa = 3.93GCGTEE199 pKa = 4.57FVTAHH204 pKa = 6.26GAFSYY209 pKa = 10.31FAEE212 pKa = 4.7EE213 pKa = 4.23YY214 pKa = 11.07GLVQHH219 pKa = 7.12AIIAGGDD226 pKa = 3.49PHH228 pKa = 8.74GEE230 pKa = 3.95ASPHH234 pKa = 7.08DD235 pKa = 3.97IGDD238 pKa = 4.14AAAHH242 pKa = 5.84AAEE245 pKa = 4.32TGYY248 pKa = 10.51ILAEE252 pKa = 4.25SPSRR256 pKa = 11.84LVDD259 pKa = 3.38ATAAEE264 pKa = 4.4AGAEE268 pKa = 4.04VLILDD273 pKa = 4.38PLEE276 pKa = 4.41TSEE279 pKa = 5.41GPYY282 pKa = 9.78EE283 pKa = 4.07DD284 pKa = 3.29VMRR287 pKa = 11.84RR288 pKa = 11.84NLGVLQQALCTQQ300 pKa = 3.87
Molecular weight: 31.22 kDa
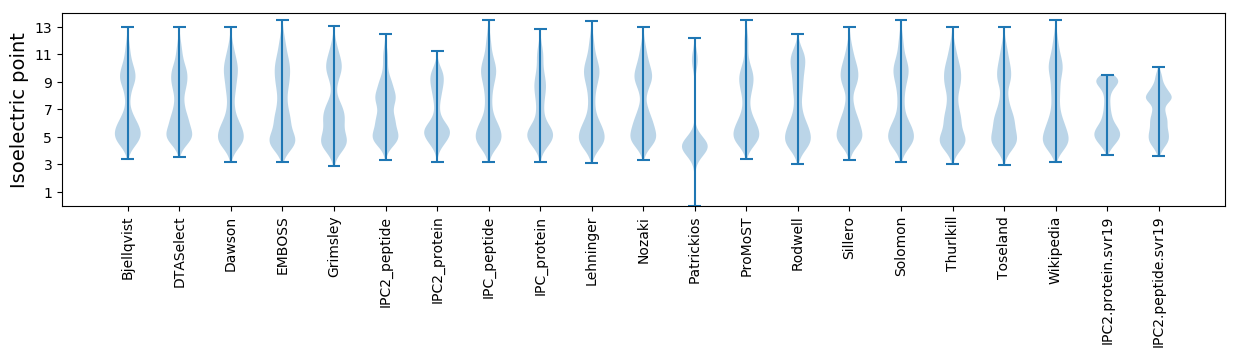
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A447G2F6|A0A447G2F6_9ARCH 30S ribosomal protein S4 OS=Nitrosopumilaceae archaeon OX=2202732 GN=rps4 PE=3 SV=1
MM1 pKa = 7.37MMMQRR6 pKa = 11.84TWRR9 pKa = 11.84RR10 pKa = 11.84PAPRR14 pKa = 11.84MAPRR18 pKa = 11.84RR19 pKa = 11.84VRR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84NRR26 pKa = 11.84PRR28 pKa = 11.84QVTLRR33 pKa = 11.84PRR35 pKa = 11.84NRR37 pKa = 11.84PGQAARR43 pKa = 11.84QLRR46 pKa = 11.84NRR48 pKa = 11.84PRR50 pKa = 11.84QVTLRR55 pKa = 11.84PRR57 pKa = 11.84NRR59 pKa = 11.84PGQAARR65 pKa = 11.84QLRR68 pKa = 11.84NRR70 pKa = 11.84PRR72 pKa = 11.84RR73 pKa = 11.84PRR75 pKa = 11.84SRR77 pKa = 11.84HH78 pKa = 5.36RR79 pKa = 11.84PAQIPPGSRR88 pKa = 11.84RR89 pKa = 11.84RR90 pKa = 11.84QTAPPPEE97 pKa = 4.59GLPCAPP103 pKa = 4.65
MM1 pKa = 7.37MMMQRR6 pKa = 11.84TWRR9 pKa = 11.84RR10 pKa = 11.84PAPRR14 pKa = 11.84MAPRR18 pKa = 11.84RR19 pKa = 11.84VRR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84NRR26 pKa = 11.84PRR28 pKa = 11.84QVTLRR33 pKa = 11.84PRR35 pKa = 11.84NRR37 pKa = 11.84PGQAARR43 pKa = 11.84QLRR46 pKa = 11.84NRR48 pKa = 11.84PRR50 pKa = 11.84QVTLRR55 pKa = 11.84PRR57 pKa = 11.84NRR59 pKa = 11.84PGQAARR65 pKa = 11.84QLRR68 pKa = 11.84NRR70 pKa = 11.84PRR72 pKa = 11.84RR73 pKa = 11.84PRR75 pKa = 11.84SRR77 pKa = 11.84HH78 pKa = 5.36RR79 pKa = 11.84PAQIPPGSRR88 pKa = 11.84RR89 pKa = 11.84RR90 pKa = 11.84QTAPPPEE97 pKa = 4.59GLPCAPP103 pKa = 4.65
Molecular weight: 12.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
579143 |
20 |
5955 |
258.9 |
28.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.091 ± 0.094 | 1.268 ± 0.027 |
6.231 ± 0.069 | 6.332 ± 0.045 |
3.078 ± 0.033 | 10.251 ± 0.077 |
2.056 ± 0.024 | 5.152 ± 0.044 |
3.448 ± 0.072 | 8.242 ± 0.072 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.935 ± 0.039 | 2.26 ± 0.04 |
5.292 ± 0.062 | 2.043 ± 0.029 |
7.978 ± 0.074 | 6.236 ± 0.039 |
4.72 ± 0.087 | 7.421 ± 0.056 |
1.025 ± 0.022 | 2.942 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
