
Pseudogymnoascus sp. VKM F-4246
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Leotiomycetes; Leotiomycetes incertae sedis; Pseudeurotiaceae; Pseudogymnoascus; unclassified Pseudogymnoascus
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
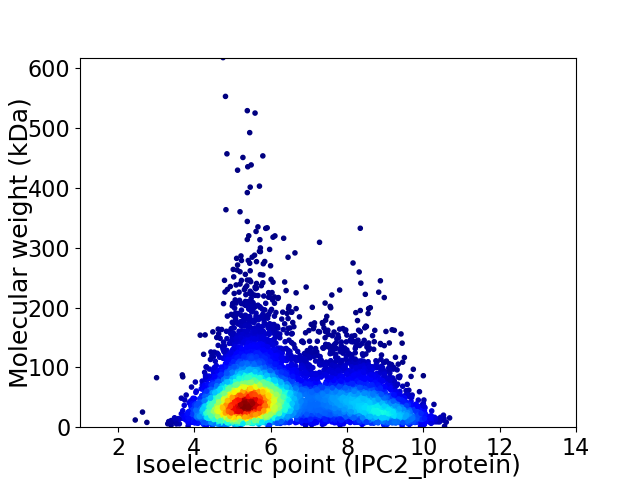
Virtual 2D-PAGE plot for 8539 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

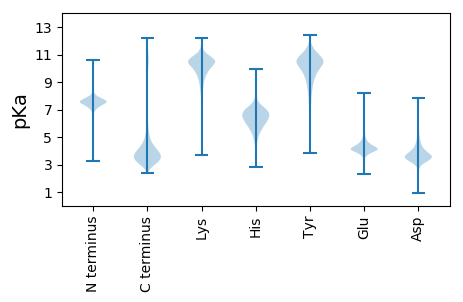
Protein with the lowest isoelectric point:
>tr|A0A093YUM7|A0A093YUM7_9PEZI Uncharacterized protein (Fragment) OS=Pseudogymnoascus sp. VKM F-4246 OX=1420902 GN=V492_05966 PE=4 SV=1
MM1 pKa = 7.62RR2 pKa = 11.84SSVLVLASVVGVALAQKK19 pKa = 10.75ASDD22 pKa = 3.87CSDD25 pKa = 3.41PQKK28 pKa = 11.24AEE30 pKa = 3.98SCVLKK35 pKa = 9.47LTKK38 pKa = 10.38AGLSCQTVLGIKK50 pKa = 9.54SCYY53 pKa = 10.36DD54 pKa = 3.43DD55 pKa = 4.34FCPEE59 pKa = 3.74IPLPDD64 pKa = 4.83EE65 pKa = 4.26INSIIDD71 pKa = 3.27TCEE74 pKa = 3.7NGGTFPGSGSGSGSGSGSGSGSGTGSGTGSEE105 pKa = 4.46SGTTTSSASQSTSSTNIPSDD125 pKa = 3.59CTDD128 pKa = 3.57ADD130 pKa = 3.9GFSDD134 pKa = 4.36CVKK137 pKa = 10.31PLVKK141 pKa = 10.24VDD143 pKa = 3.79TTCKK147 pKa = 9.37VTMQIQQCFKK157 pKa = 9.64TYY159 pKa = 10.16CPQHH163 pKa = 6.58IDD165 pKa = 3.8RR166 pKa = 11.84IEE168 pKa = 5.1DD169 pKa = 3.75DD170 pKa = 3.71VTSCQGSEE178 pKa = 4.25DD179 pKa = 3.85GSGTGSDD186 pKa = 4.55DD187 pKa = 4.52DD188 pKa = 4.09STPTSSSDD196 pKa = 3.27DD197 pKa = 3.78SEE199 pKa = 4.55GTGISGGGSSTDD211 pKa = 3.28TSSSADD217 pKa = 3.43STGTGVDD224 pKa = 3.26SGSDD228 pKa = 3.53NTSSSSDD235 pKa = 3.35PTNTSTNSDD244 pKa = 3.52PVATDD249 pKa = 3.32SNVNEE254 pKa = 4.29GAGDD258 pKa = 3.84KK259 pKa = 10.9LFAPAGAIIGSIIAVMAWLL278 pKa = 3.98
MM1 pKa = 7.62RR2 pKa = 11.84SSVLVLASVVGVALAQKK19 pKa = 10.75ASDD22 pKa = 3.87CSDD25 pKa = 3.41PQKK28 pKa = 11.24AEE30 pKa = 3.98SCVLKK35 pKa = 9.47LTKK38 pKa = 10.38AGLSCQTVLGIKK50 pKa = 9.54SCYY53 pKa = 10.36DD54 pKa = 3.43DD55 pKa = 4.34FCPEE59 pKa = 3.74IPLPDD64 pKa = 4.83EE65 pKa = 4.26INSIIDD71 pKa = 3.27TCEE74 pKa = 3.7NGGTFPGSGSGSGSGSGSGSGSGTGSGTGSEE105 pKa = 4.46SGTTTSSASQSTSSTNIPSDD125 pKa = 3.59CTDD128 pKa = 3.57ADD130 pKa = 3.9GFSDD134 pKa = 4.36CVKK137 pKa = 10.31PLVKK141 pKa = 10.24VDD143 pKa = 3.79TTCKK147 pKa = 9.37VTMQIQQCFKK157 pKa = 9.64TYY159 pKa = 10.16CPQHH163 pKa = 6.58IDD165 pKa = 3.8RR166 pKa = 11.84IEE168 pKa = 5.1DD169 pKa = 3.75DD170 pKa = 3.71VTSCQGSEE178 pKa = 4.25DD179 pKa = 3.85GSGTGSDD186 pKa = 4.55DD187 pKa = 4.52DD188 pKa = 4.09STPTSSSDD196 pKa = 3.27DD197 pKa = 3.78SEE199 pKa = 4.55GTGISGGGSSTDD211 pKa = 3.28TSSSADD217 pKa = 3.43STGTGVDD224 pKa = 3.26SGSDD228 pKa = 3.53NTSSSSDD235 pKa = 3.35PTNTSTNSDD244 pKa = 3.52PVATDD249 pKa = 3.32SNVNEE254 pKa = 4.29GAGDD258 pKa = 3.84KK259 pKa = 10.9LFAPAGAIIGSIIAVMAWLL278 pKa = 3.98
Molecular weight: 27.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A093YLY4|A0A093YLY4_9PEZI Acylphosphatase OS=Pseudogymnoascus sp. VKM F-4246 OX=1420902 GN=V492_08180 PE=3 SV=1
MM1 pKa = 7.44VSKK4 pKa = 10.83LPRR7 pKa = 11.84NTTLLGSPSEE17 pKa = 4.17NNRR20 pKa = 11.84NLVIVLSAVFGSLALFMVVSATVFVYY46 pKa = 9.94RR47 pKa = 11.84WKK49 pKa = 10.77RR50 pKa = 11.84NPKK53 pKa = 6.65YY54 pKa = 9.72HH55 pKa = 6.87RR56 pKa = 11.84RR57 pKa = 11.84GISPVDD63 pKa = 3.43DD64 pKa = 4.09EE65 pKa = 5.41EE66 pKa = 4.64IEE68 pKa = 3.92RR69 pKa = 11.84WRR71 pKa = 11.84GRR73 pKa = 11.84KK74 pKa = 7.09EE75 pKa = 4.11TYY77 pKa = 9.7TEE79 pKa = 4.34APSRR83 pKa = 11.84TAPKK87 pKa = 7.38THH89 pKa = 6.54KK90 pKa = 9.33RR91 pKa = 11.84QSSSVVIVSNPPGWTWSAEE110 pKa = 3.67PSPFGTRR117 pKa = 11.84NSGEE121 pKa = 4.34TALSPPPMVARR132 pKa = 11.84APNSRR137 pKa = 11.84SGLTDD142 pKa = 3.5GAILGADD149 pKa = 3.35PFVPPVRR156 pKa = 11.84RR157 pKa = 11.84PSARR161 pKa = 11.84LAKK164 pKa = 10.15HH165 pKa = 6.01SRR167 pKa = 11.84NQSRR171 pKa = 11.84KK172 pKa = 8.11SSFSASIPEE181 pKa = 4.27RR182 pKa = 11.84MSTDD186 pKa = 3.27LPRR189 pKa = 11.84HH190 pKa = 5.45YY191 pKa = 10.65RR192 pKa = 11.84DD193 pKa = 3.7DD194 pKa = 3.9SKK196 pKa = 11.71KK197 pKa = 10.66SFQADD202 pKa = 3.53PVSPPSSIFNGSPGTNEE219 pKa = 4.23PLPTSDD225 pKa = 3.97TGFCRR230 pKa = 11.84LPSRR234 pKa = 11.84SIDD237 pKa = 3.61SLLGPLARR245 pKa = 11.84PLPNPPLHH253 pKa = 6.56QLDD256 pKa = 4.76AVDD259 pKa = 5.26APPPPPPLPHH269 pKa = 6.9PTQQPRR275 pKa = 11.84ARR277 pKa = 11.84LPRR280 pKa = 11.84RR281 pKa = 11.84RR282 pKa = 11.84IPRR285 pKa = 11.84RR286 pKa = 11.84AARR289 pKa = 11.84DD290 pKa = 4.13AIPLDD295 pKa = 3.78ARR297 pKa = 11.84DD298 pKa = 4.16GVHH301 pKa = 6.84AAHH304 pKa = 6.41GCVWGAAYY312 pKa = 10.18LWAGVLWRR320 pKa = 11.84DD321 pKa = 3.41CGVWNDD327 pKa = 3.67GVSEE331 pKa = 4.02GRR333 pKa = 11.84GEE335 pKa = 4.23DD336 pKa = 3.48GKK338 pKa = 11.23
MM1 pKa = 7.44VSKK4 pKa = 10.83LPRR7 pKa = 11.84NTTLLGSPSEE17 pKa = 4.17NNRR20 pKa = 11.84NLVIVLSAVFGSLALFMVVSATVFVYY46 pKa = 9.94RR47 pKa = 11.84WKK49 pKa = 10.77RR50 pKa = 11.84NPKK53 pKa = 6.65YY54 pKa = 9.72HH55 pKa = 6.87RR56 pKa = 11.84RR57 pKa = 11.84GISPVDD63 pKa = 3.43DD64 pKa = 4.09EE65 pKa = 5.41EE66 pKa = 4.64IEE68 pKa = 3.92RR69 pKa = 11.84WRR71 pKa = 11.84GRR73 pKa = 11.84KK74 pKa = 7.09EE75 pKa = 4.11TYY77 pKa = 9.7TEE79 pKa = 4.34APSRR83 pKa = 11.84TAPKK87 pKa = 7.38THH89 pKa = 6.54KK90 pKa = 9.33RR91 pKa = 11.84QSSSVVIVSNPPGWTWSAEE110 pKa = 3.67PSPFGTRR117 pKa = 11.84NSGEE121 pKa = 4.34TALSPPPMVARR132 pKa = 11.84APNSRR137 pKa = 11.84SGLTDD142 pKa = 3.5GAILGADD149 pKa = 3.35PFVPPVRR156 pKa = 11.84RR157 pKa = 11.84PSARR161 pKa = 11.84LAKK164 pKa = 10.15HH165 pKa = 6.01SRR167 pKa = 11.84NQSRR171 pKa = 11.84KK172 pKa = 8.11SSFSASIPEE181 pKa = 4.27RR182 pKa = 11.84MSTDD186 pKa = 3.27LPRR189 pKa = 11.84HH190 pKa = 5.45YY191 pKa = 10.65RR192 pKa = 11.84DD193 pKa = 3.7DD194 pKa = 3.9SKK196 pKa = 11.71KK197 pKa = 10.66SFQADD202 pKa = 3.53PVSPPSSIFNGSPGTNEE219 pKa = 4.23PLPTSDD225 pKa = 3.97TGFCRR230 pKa = 11.84LPSRR234 pKa = 11.84SIDD237 pKa = 3.61SLLGPLARR245 pKa = 11.84PLPNPPLHH253 pKa = 6.56QLDD256 pKa = 4.76AVDD259 pKa = 5.26APPPPPPLPHH269 pKa = 6.9PTQQPRR275 pKa = 11.84ARR277 pKa = 11.84LPRR280 pKa = 11.84RR281 pKa = 11.84RR282 pKa = 11.84IPRR285 pKa = 11.84RR286 pKa = 11.84AARR289 pKa = 11.84DD290 pKa = 4.13AIPLDD295 pKa = 3.78ARR297 pKa = 11.84DD298 pKa = 4.16GVHH301 pKa = 6.84AAHH304 pKa = 6.41GCVWGAAYY312 pKa = 10.18LWAGVLWRR320 pKa = 11.84DD321 pKa = 3.41CGVWNDD327 pKa = 3.67GVSEE331 pKa = 4.02GRR333 pKa = 11.84GEE335 pKa = 4.23DD336 pKa = 3.48GKK338 pKa = 11.23
Molecular weight: 37.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4315648 |
10 |
5650 |
505.4 |
55.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.947 ± 0.024 | 1.157 ± 0.01 |
5.761 ± 0.02 | 6.351 ± 0.026 |
3.608 ± 0.015 | 7.275 ± 0.028 |
2.216 ± 0.011 | 5.003 ± 0.018 |
5.081 ± 0.02 | 8.658 ± 0.028 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.155 ± 0.01 | 3.765 ± 0.013 |
5.915 ± 0.025 | 3.861 ± 0.019 |
5.784 ± 0.02 | 8.097 ± 0.026 |
6.016 ± 0.016 | 6.176 ± 0.018 |
1.407 ± 0.009 | 2.766 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
