
Petromyzon marinus (Sea lamprey)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Cyclostomata; Hyperoartia; Petromyzontiformes; Petromyzontidae; Petromyzon
Average proteome isoelectric point is 6.78
Get precalculated fractions of proteins
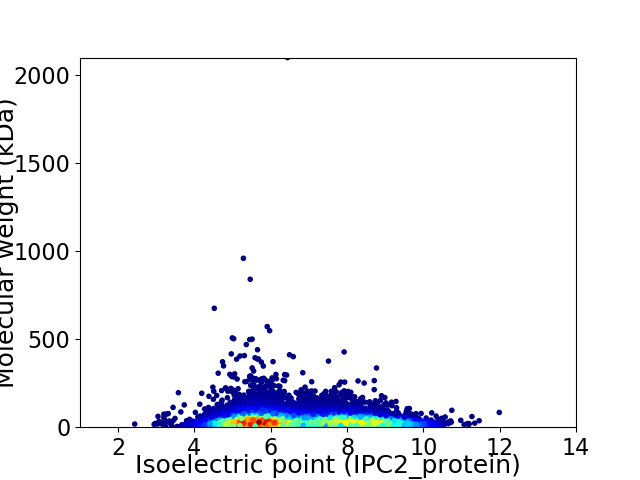
Virtual 2D-PAGE plot for 11407 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|S4RPW3|S4RPW3_PETMA Isoform of S4RPV9 BRF1 RNA polymerase III transcription initiation factor subunit OS=Petromyzon marinus OX=7757 PE=4 SV=1
TT1 pKa = 7.43ALLGNDD7 pKa = 3.15NVYY10 pKa = 10.78EE11 pKa = 4.16LNLDD15 pKa = 3.86YY16 pKa = 11.39NQLQTLSAGVFDD28 pKa = 6.15DD29 pKa = 3.79LTEE32 pKa = 4.7LGTLGLANNQLASLPLGVFADD53 pKa = 4.79LAPTDD58 pKa = 4.62DD59 pKa = 5.16LYY61 pKa = 12.08LNINQLKK68 pKa = 9.37SLPSGVFDD76 pKa = 4.87SLTKK80 pKa = 9.99LTILQLDD87 pKa = 4.13YY88 pKa = 11.43NQLQSVPHH96 pKa = 6.1GAFDD100 pKa = 5.0RR101 pKa = 11.84LTNLQTMQLHH111 pKa = 6.26SDD113 pKa = 3.84VLQSVPNGAFNALTKK128 pKa = 10.88LEE130 pKa = 4.56TITLNVNVWDD140 pKa = 4.88CSSCDD145 pKa = 3.21ILYY148 pKa = 10.69LSEE151 pKa = 4.02
TT1 pKa = 7.43ALLGNDD7 pKa = 3.15NVYY10 pKa = 10.78EE11 pKa = 4.16LNLDD15 pKa = 3.86YY16 pKa = 11.39NQLQTLSAGVFDD28 pKa = 6.15DD29 pKa = 3.79LTEE32 pKa = 4.7LGTLGLANNQLASLPLGVFADD53 pKa = 4.79LAPTDD58 pKa = 4.62DD59 pKa = 5.16LYY61 pKa = 12.08LNINQLKK68 pKa = 9.37SLPSGVFDD76 pKa = 4.87SLTKK80 pKa = 9.99LTILQLDD87 pKa = 4.13YY88 pKa = 11.43NQLQSVPHH96 pKa = 6.1GAFDD100 pKa = 5.0RR101 pKa = 11.84LTNLQTMQLHH111 pKa = 6.26SDD113 pKa = 3.84VLQSVPNGAFNALTKK128 pKa = 10.88LEE130 pKa = 4.56TITLNVNVWDD140 pKa = 4.88CSSCDD145 pKa = 3.21ILYY148 pKa = 10.69LSEE151 pKa = 4.02
Molecular weight: 16.52 kDa
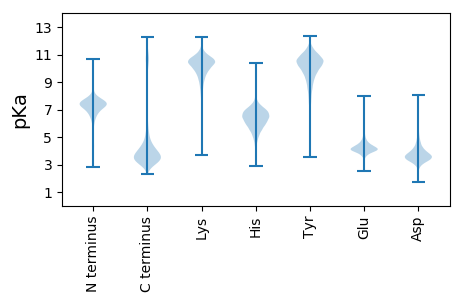
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S4RWB8|S4RWB8_PETMA RNA-binding motif protein 42 OS=Petromyzon marinus OX=7757 PE=3 SV=1
PP1 pKa = 7.3SPLDD5 pKa = 3.43SSEE8 pKa = 3.94AATAQRR14 pKa = 11.84AKK16 pKa = 10.45FADD19 pKa = 3.92SSSSTLFTSMAALNHH34 pKa = 6.51YY35 pKa = 7.63DD36 pKa = 3.53THH38 pKa = 7.6CNGYY42 pKa = 8.44HH43 pKa = 6.46CKK45 pKa = 10.2HH46 pKa = 5.26VVRR49 pKa = 11.84LISRR53 pKa = 11.84HH54 pKa = 5.18LIARR58 pKa = 11.84KK59 pKa = 9.57VNVVRR64 pKa = 11.84LIIRR68 pKa = 11.84HH69 pKa = 5.17LNARR73 pKa = 11.84KK74 pKa = 9.83VNVVRR79 pKa = 11.84LIIRR83 pKa = 11.84HH84 pKa = 5.91LIARR88 pKa = 11.84KK89 pKa = 9.27VNVVRR94 pKa = 11.84LISRR98 pKa = 11.84HH99 pKa = 5.18LIARR103 pKa = 11.84KK104 pKa = 9.57VNVVRR109 pKa = 11.84LIIRR113 pKa = 11.84HH114 pKa = 5.91LIARR118 pKa = 11.84KK119 pKa = 9.27VNVVRR124 pKa = 11.84LIIRR128 pKa = 11.84HH129 pKa = 5.91LIARR133 pKa = 11.84KK134 pKa = 9.37VNMVRR139 pKa = 11.84LISRR143 pKa = 11.84HH144 pKa = 5.17LIARR148 pKa = 11.84KK149 pKa = 9.57VNVVRR154 pKa = 11.84LIIRR158 pKa = 11.84HH159 pKa = 5.17LNARR163 pKa = 11.84KK164 pKa = 9.83VNVVRR169 pKa = 11.84LIIRR173 pKa = 11.84HH174 pKa = 5.24LNARR178 pKa = 11.84KK179 pKa = 9.21HH180 pKa = 5.65LIARR184 pKa = 11.84KK185 pKa = 8.92VNVVRR190 pKa = 11.84LIIRR194 pKa = 11.84HH195 pKa = 5.17LNARR199 pKa = 11.84KK200 pKa = 9.83VNVVRR205 pKa = 11.84LIIRR209 pKa = 11.84HH210 pKa = 5.91LIARR214 pKa = 11.84KK215 pKa = 9.27VNVVRR220 pKa = 11.84LIIRR224 pKa = 11.84HH225 pKa = 5.91LIARR229 pKa = 11.84KK230 pKa = 9.27VNVVRR235 pKa = 11.84LIIRR239 pKa = 11.84HH240 pKa = 5.55LNARR244 pKa = 11.84HH245 pKa = 5.07LNARR249 pKa = 11.84KK250 pKa = 10.39VNVHH254 pKa = 6.53LIARR258 pKa = 11.84KK259 pKa = 9.14VNVVRR264 pKa = 11.84LIIRR268 pKa = 11.84HH269 pKa = 5.17LNARR273 pKa = 11.84KK274 pKa = 9.83VNVVRR279 pKa = 11.84LIILHH284 pKa = 6.41LIARR288 pKa = 11.84KK289 pKa = 9.81VNVVRR294 pKa = 11.84LIIRR298 pKa = 11.84HH299 pKa = 5.91LIARR303 pKa = 11.84KK304 pKa = 9.27VNVVRR309 pKa = 11.84LIIRR313 pKa = 11.84HH314 pKa = 5.17LNARR318 pKa = 11.84KK319 pKa = 10.12VNVVHH324 pKa = 7.05LIIRR328 pKa = 11.84HH329 pKa = 5.63LIARR333 pKa = 11.84KK334 pKa = 9.27VNVVRR339 pKa = 11.84LIIQHH344 pKa = 6.61LIARR348 pKa = 11.84KK349 pKa = 8.99VNVVRR354 pKa = 11.84LIIRR358 pKa = 11.84HH359 pKa = 5.17LNARR363 pKa = 11.84KK364 pKa = 9.83VNVVRR369 pKa = 11.84LIIRR373 pKa = 11.84HH374 pKa = 5.91LIARR378 pKa = 11.84KK379 pKa = 9.27VNVVRR384 pKa = 11.84LIIRR388 pKa = 11.84HH389 pKa = 5.91LIARR393 pKa = 11.84KK394 pKa = 9.27VNVVRR399 pKa = 11.84LIIRR403 pKa = 11.84HH404 pKa = 5.91LIARR408 pKa = 11.84KK409 pKa = 9.27VNVVRR414 pKa = 11.84LIIRR418 pKa = 11.84HH419 pKa = 5.17LNARR423 pKa = 11.84KK424 pKa = 9.83VNVVRR429 pKa = 11.84LIIRR433 pKa = 11.84HH434 pKa = 5.29LNARR438 pKa = 11.84KK439 pKa = 9.99ANVVRR444 pKa = 11.84LIIRR448 pKa = 11.84HH449 pKa = 5.17LNARR453 pKa = 11.84KK454 pKa = 9.83VNVVRR459 pKa = 11.84LIIRR463 pKa = 11.84HH464 pKa = 5.91LIARR468 pKa = 11.84KK469 pKa = 9.27VNVVRR474 pKa = 11.84LIIRR478 pKa = 11.84HH479 pKa = 5.91LIARR483 pKa = 11.84KK484 pKa = 9.27VNVVRR489 pKa = 11.84LIIRR493 pKa = 11.84HH494 pKa = 5.91LIARR498 pKa = 11.84KK499 pKa = 9.27VNVVRR504 pKa = 11.84LIIRR508 pKa = 11.84HH509 pKa = 5.17LNARR513 pKa = 11.84KK514 pKa = 10.36VNVV517 pKa = 3.29
PP1 pKa = 7.3SPLDD5 pKa = 3.43SSEE8 pKa = 3.94AATAQRR14 pKa = 11.84AKK16 pKa = 10.45FADD19 pKa = 3.92SSSSTLFTSMAALNHH34 pKa = 6.51YY35 pKa = 7.63DD36 pKa = 3.53THH38 pKa = 7.6CNGYY42 pKa = 8.44HH43 pKa = 6.46CKK45 pKa = 10.2HH46 pKa = 5.26VVRR49 pKa = 11.84LISRR53 pKa = 11.84HH54 pKa = 5.18LIARR58 pKa = 11.84KK59 pKa = 9.57VNVVRR64 pKa = 11.84LIIRR68 pKa = 11.84HH69 pKa = 5.17LNARR73 pKa = 11.84KK74 pKa = 9.83VNVVRR79 pKa = 11.84LIIRR83 pKa = 11.84HH84 pKa = 5.91LIARR88 pKa = 11.84KK89 pKa = 9.27VNVVRR94 pKa = 11.84LISRR98 pKa = 11.84HH99 pKa = 5.18LIARR103 pKa = 11.84KK104 pKa = 9.57VNVVRR109 pKa = 11.84LIIRR113 pKa = 11.84HH114 pKa = 5.91LIARR118 pKa = 11.84KK119 pKa = 9.27VNVVRR124 pKa = 11.84LIIRR128 pKa = 11.84HH129 pKa = 5.91LIARR133 pKa = 11.84KK134 pKa = 9.37VNMVRR139 pKa = 11.84LISRR143 pKa = 11.84HH144 pKa = 5.17LIARR148 pKa = 11.84KK149 pKa = 9.57VNVVRR154 pKa = 11.84LIIRR158 pKa = 11.84HH159 pKa = 5.17LNARR163 pKa = 11.84KK164 pKa = 9.83VNVVRR169 pKa = 11.84LIIRR173 pKa = 11.84HH174 pKa = 5.24LNARR178 pKa = 11.84KK179 pKa = 9.21HH180 pKa = 5.65LIARR184 pKa = 11.84KK185 pKa = 8.92VNVVRR190 pKa = 11.84LIIRR194 pKa = 11.84HH195 pKa = 5.17LNARR199 pKa = 11.84KK200 pKa = 9.83VNVVRR205 pKa = 11.84LIIRR209 pKa = 11.84HH210 pKa = 5.91LIARR214 pKa = 11.84KK215 pKa = 9.27VNVVRR220 pKa = 11.84LIIRR224 pKa = 11.84HH225 pKa = 5.91LIARR229 pKa = 11.84KK230 pKa = 9.27VNVVRR235 pKa = 11.84LIIRR239 pKa = 11.84HH240 pKa = 5.55LNARR244 pKa = 11.84HH245 pKa = 5.07LNARR249 pKa = 11.84KK250 pKa = 10.39VNVHH254 pKa = 6.53LIARR258 pKa = 11.84KK259 pKa = 9.14VNVVRR264 pKa = 11.84LIIRR268 pKa = 11.84HH269 pKa = 5.17LNARR273 pKa = 11.84KK274 pKa = 9.83VNVVRR279 pKa = 11.84LIILHH284 pKa = 6.41LIARR288 pKa = 11.84KK289 pKa = 9.81VNVVRR294 pKa = 11.84LIIRR298 pKa = 11.84HH299 pKa = 5.91LIARR303 pKa = 11.84KK304 pKa = 9.27VNVVRR309 pKa = 11.84LIIRR313 pKa = 11.84HH314 pKa = 5.17LNARR318 pKa = 11.84KK319 pKa = 10.12VNVVHH324 pKa = 7.05LIIRR328 pKa = 11.84HH329 pKa = 5.63LIARR333 pKa = 11.84KK334 pKa = 9.27VNVVRR339 pKa = 11.84LIIQHH344 pKa = 6.61LIARR348 pKa = 11.84KK349 pKa = 8.99VNVVRR354 pKa = 11.84LIIRR358 pKa = 11.84HH359 pKa = 5.17LNARR363 pKa = 11.84KK364 pKa = 9.83VNVVRR369 pKa = 11.84LIIRR373 pKa = 11.84HH374 pKa = 5.91LIARR378 pKa = 11.84KK379 pKa = 9.27VNVVRR384 pKa = 11.84LIIRR388 pKa = 11.84HH389 pKa = 5.91LIARR393 pKa = 11.84KK394 pKa = 9.27VNVVRR399 pKa = 11.84LIIRR403 pKa = 11.84HH404 pKa = 5.91LIARR408 pKa = 11.84KK409 pKa = 9.27VNVVRR414 pKa = 11.84LIIRR418 pKa = 11.84HH419 pKa = 5.17LNARR423 pKa = 11.84KK424 pKa = 9.83VNVVRR429 pKa = 11.84LIIRR433 pKa = 11.84HH434 pKa = 5.29LNARR438 pKa = 11.84KK439 pKa = 9.99ANVVRR444 pKa = 11.84LIIRR448 pKa = 11.84HH449 pKa = 5.17LNARR453 pKa = 11.84KK454 pKa = 9.83VNVVRR459 pKa = 11.84LIIRR463 pKa = 11.84HH464 pKa = 5.91LIARR468 pKa = 11.84KK469 pKa = 9.27VNVVRR474 pKa = 11.84LIIRR478 pKa = 11.84HH479 pKa = 5.91LIARR483 pKa = 11.84KK484 pKa = 9.27VNVVRR489 pKa = 11.84LIIRR493 pKa = 11.84HH494 pKa = 5.91LIARR498 pKa = 11.84KK499 pKa = 9.27VNVVRR504 pKa = 11.84LIIRR508 pKa = 11.84HH509 pKa = 5.17LNARR513 pKa = 11.84KK514 pKa = 10.36VNVV517 pKa = 3.29
Molecular weight: 60.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
4607059 |
9 |
18900 |
403.9 |
44.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.042 ± 0.029 | 2.376 ± 0.025 |
5.078 ± 0.019 | 6.47 ± 0.032 |
3.634 ± 0.018 | 7.049 ± 0.034 |
2.772 ± 0.014 | 3.971 ± 0.02 |
4.81 ± 0.031 | 9.97 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.235 ± 0.012 | 3.419 ± 0.019 |
5.557 ± 0.032 | 4.355 ± 0.025 |
6.482 ± 0.025 | 7.628 ± 0.026 |
5.287 ± 0.024 | 6.914 ± 0.021 |
1.28 ± 0.01 | 2.666 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
