
Mesorhizobium albiziae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Phyllobacteriaceae; Mesorhizobium
Average proteome isoelectric point is 6.62
Get precalculated fractions of proteins
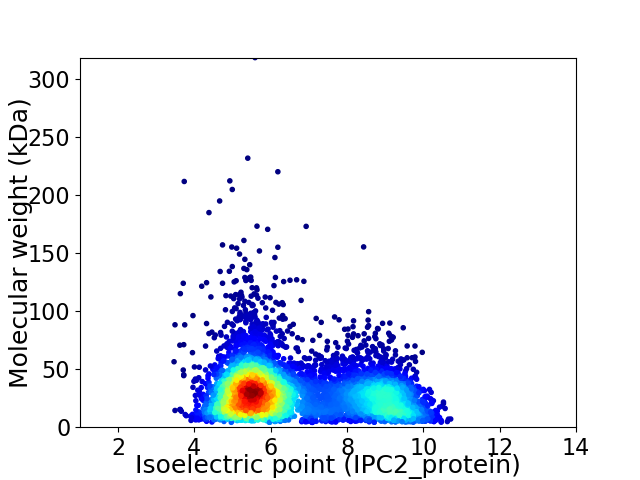
Virtual 2D-PAGE plot for 6112 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I4EUV0|A0A1I4EUV0_9RHIZ DNA-binding transcriptional regulator LysR family OS=Mesorhizobium albiziae OX=335020 GN=SAMN04488498_13034 PE=3 SV=1
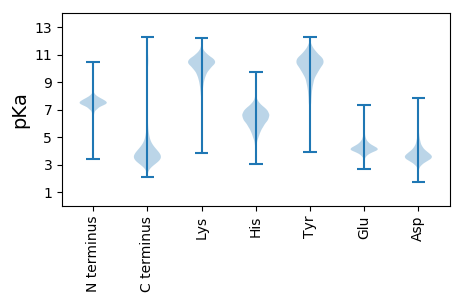
MM1 pKa = 7.93DD2 pKa = 5.32FKK4 pKa = 11.57SLLLGSAAALVAVSAARR21 pKa = 11.84AADD24 pKa = 3.19AVVIPEE30 pKa = 4.5PEE32 pKa = 3.71PVEE35 pKa = 4.05YY36 pKa = 11.08VRR38 pKa = 11.84VCDD41 pKa = 3.84VYY43 pKa = 10.71GTAFYY48 pKa = 9.86YY49 pKa = 10.19IPGTEE54 pKa = 3.92TCLKK58 pKa = 8.47VSGYY62 pKa = 9.13VRR64 pKa = 11.84YY65 pKa = 10.19DD66 pKa = 2.94IGVGDD71 pKa = 4.45LFGLQSADD79 pKa = 3.46KK80 pKa = 10.68LDD82 pKa = 3.86LVEE85 pKa = 6.94DD86 pKa = 4.29GVLDD90 pKa = 4.54INDD93 pKa = 4.01TYY95 pKa = 11.13FKK97 pKa = 10.4RR98 pKa = 11.84ARR100 pKa = 11.84AALRR104 pKa = 11.84LDD106 pKa = 3.2ARR108 pKa = 11.84TEE110 pKa = 4.27TEE112 pKa = 4.07LGTLRR117 pKa = 11.84AYY119 pKa = 9.64TEE121 pKa = 5.88LKK123 pKa = 10.02FQWDD127 pKa = 4.1TNQDD131 pKa = 3.45SVDD134 pKa = 3.73VVDD137 pKa = 6.27PIAGDD142 pKa = 3.69TVTVPFVSAGNEE154 pKa = 3.67FAMEE158 pKa = 3.93DD159 pKa = 3.91VYY161 pKa = 11.29IEE163 pKa = 4.15LGGFRR168 pKa = 11.84IGATDD173 pKa = 3.46SLFTSFTDD181 pKa = 3.57YY182 pKa = 11.42AGAVINDD189 pKa = 4.33DD190 pKa = 3.73FLVPYY195 pKa = 10.33GPLDD199 pKa = 3.43GDD201 pKa = 3.66MTHH204 pKa = 6.59QIAYY208 pKa = 7.94TFTADD213 pKa = 3.47NGFTAAIAIEE223 pKa = 4.5EE224 pKa = 4.6GNGDD228 pKa = 3.72PDD230 pKa = 4.11AVFGSDD236 pKa = 3.16ADD238 pKa = 3.79GDD240 pKa = 4.5GVVDD244 pKa = 3.86TVTTIEE250 pKa = 3.93AAYY253 pKa = 9.13TLDD256 pKa = 4.48DD257 pKa = 4.63YY258 pKa = 11.33IPHH261 pKa = 5.65VVAGVGYY268 pKa = 9.78AGGWGSIKK276 pKa = 10.74AVGAYY281 pKa = 9.92DD282 pKa = 4.03AVWEE286 pKa = 4.22EE287 pKa = 3.69WAAKK291 pKa = 9.8VRR293 pKa = 11.84LDD295 pKa = 3.62VKK297 pKa = 10.14ATQTISAFIMAGYY310 pKa = 9.82KK311 pKa = 10.11SSEE314 pKa = 4.45DD315 pKa = 3.78FSITDD320 pKa = 3.71PVTGITTTFDD330 pKa = 3.19GPNYY334 pKa = 8.05YY335 pKa = 10.84GNWGGEE341 pKa = 3.67WAIWGGGSVEE351 pKa = 4.25VTEE354 pKa = 4.28KK355 pKa = 10.62ATFNVQLAYY364 pKa = 10.67DD365 pKa = 3.81DD366 pKa = 4.77FEE368 pKa = 4.79NFSAVANVAYY378 pKa = 9.7EE379 pKa = 4.05VVPGFMITPEE389 pKa = 3.59VAYY392 pKa = 10.32ADD394 pKa = 3.75NFEE397 pKa = 5.84DD398 pKa = 5.15SDD400 pKa = 5.01LDD402 pKa = 4.27DD403 pKa = 4.99FDD405 pKa = 6.41GEE407 pKa = 4.24FGGYY411 pKa = 10.05LRR413 pKa = 11.84FQRR416 pKa = 11.84NFF418 pKa = 3.01
MM1 pKa = 7.93DD2 pKa = 5.32FKK4 pKa = 11.57SLLLGSAAALVAVSAARR21 pKa = 11.84AADD24 pKa = 3.19AVVIPEE30 pKa = 4.5PEE32 pKa = 3.71PVEE35 pKa = 4.05YY36 pKa = 11.08VRR38 pKa = 11.84VCDD41 pKa = 3.84VYY43 pKa = 10.71GTAFYY48 pKa = 9.86YY49 pKa = 10.19IPGTEE54 pKa = 3.92TCLKK58 pKa = 8.47VSGYY62 pKa = 9.13VRR64 pKa = 11.84YY65 pKa = 10.19DD66 pKa = 2.94IGVGDD71 pKa = 4.45LFGLQSADD79 pKa = 3.46KK80 pKa = 10.68LDD82 pKa = 3.86LVEE85 pKa = 6.94DD86 pKa = 4.29GVLDD90 pKa = 4.54INDD93 pKa = 4.01TYY95 pKa = 11.13FKK97 pKa = 10.4RR98 pKa = 11.84ARR100 pKa = 11.84AALRR104 pKa = 11.84LDD106 pKa = 3.2ARR108 pKa = 11.84TEE110 pKa = 4.27TEE112 pKa = 4.07LGTLRR117 pKa = 11.84AYY119 pKa = 9.64TEE121 pKa = 5.88LKK123 pKa = 10.02FQWDD127 pKa = 4.1TNQDD131 pKa = 3.45SVDD134 pKa = 3.73VVDD137 pKa = 6.27PIAGDD142 pKa = 3.69TVTVPFVSAGNEE154 pKa = 3.67FAMEE158 pKa = 3.93DD159 pKa = 3.91VYY161 pKa = 11.29IEE163 pKa = 4.15LGGFRR168 pKa = 11.84IGATDD173 pKa = 3.46SLFTSFTDD181 pKa = 3.57YY182 pKa = 11.42AGAVINDD189 pKa = 4.33DD190 pKa = 3.73FLVPYY195 pKa = 10.33GPLDD199 pKa = 3.43GDD201 pKa = 3.66MTHH204 pKa = 6.59QIAYY208 pKa = 7.94TFTADD213 pKa = 3.47NGFTAAIAIEE223 pKa = 4.5EE224 pKa = 4.6GNGDD228 pKa = 3.72PDD230 pKa = 4.11AVFGSDD236 pKa = 3.16ADD238 pKa = 3.79GDD240 pKa = 4.5GVVDD244 pKa = 3.86TVTTIEE250 pKa = 3.93AAYY253 pKa = 9.13TLDD256 pKa = 4.48DD257 pKa = 4.63YY258 pKa = 11.33IPHH261 pKa = 5.65VVAGVGYY268 pKa = 9.78AGGWGSIKK276 pKa = 10.74AVGAYY281 pKa = 9.92DD282 pKa = 4.03AVWEE286 pKa = 4.22EE287 pKa = 3.69WAAKK291 pKa = 9.8VRR293 pKa = 11.84LDD295 pKa = 3.62VKK297 pKa = 10.14ATQTISAFIMAGYY310 pKa = 9.82KK311 pKa = 10.11SSEE314 pKa = 4.45DD315 pKa = 3.78FSITDD320 pKa = 3.71PVTGITTTFDD330 pKa = 3.19GPNYY334 pKa = 8.05YY335 pKa = 10.84GNWGGEE341 pKa = 3.67WAIWGGGSVEE351 pKa = 4.25VTEE354 pKa = 4.28KK355 pKa = 10.62ATFNVQLAYY364 pKa = 10.67DD365 pKa = 3.81DD366 pKa = 4.77FEE368 pKa = 4.79NFSAVANVAYY378 pKa = 9.7EE379 pKa = 4.05VVPGFMITPEE389 pKa = 3.59VAYY392 pKa = 10.32ADD394 pKa = 3.75NFEE397 pKa = 5.84DD398 pKa = 5.15SDD400 pKa = 5.01LDD402 pKa = 4.27DD403 pKa = 4.99FDD405 pKa = 6.41GEE407 pKa = 4.24FGGYY411 pKa = 10.05LRR413 pKa = 11.84FQRR416 pKa = 11.84NFF418 pKa = 3.01
Molecular weight: 45.16 kDa
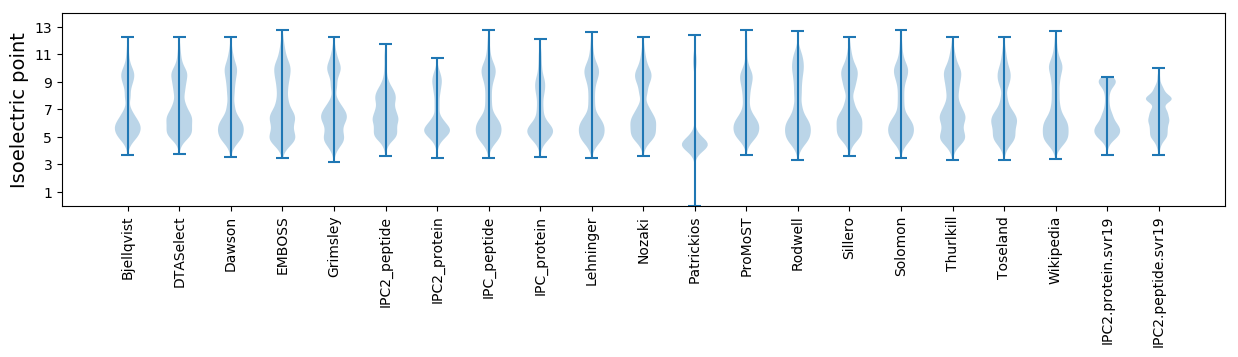
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I4FL32|A0A1I4FL32_9RHIZ Biotin carboxylase OS=Mesorhizobium albiziae OX=335020 GN=SAMN04488498_1491 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 10.47SQDD5 pKa = 3.45HH6 pKa = 6.27RR7 pKa = 11.84HH8 pKa = 5.15TAQARR13 pKa = 11.84QPTRR17 pKa = 11.84AGEE20 pKa = 4.18RR21 pKa = 11.84KK22 pKa = 9.15GCRR25 pKa = 11.84SSALGSPGRR34 pKa = 11.84RR35 pKa = 11.84SVEE38 pKa = 3.64DD39 pKa = 3.98PYY41 pKa = 11.44SVRR44 pKa = 11.84NLGSSLGISKK54 pKa = 9.06TEE56 pKa = 3.8VNASINRR63 pKa = 11.84SLASGIAARR72 pKa = 11.84DD73 pKa = 3.59RR74 pKa = 11.84DD75 pKa = 3.7TGRR78 pKa = 11.84AKK80 pKa = 10.44PSRR83 pKa = 11.84RR84 pKa = 11.84SLLNFIVHH92 pKa = 6.0GLKK95 pKa = 10.31FVFPAKK101 pKa = 9.87PGAMQRR107 pKa = 11.84GIPTAFAAPVLKK119 pKa = 10.61NALISAGSYY128 pKa = 9.73IYY130 pKa = 10.25VWPYY134 pKa = 11.59ASGHH138 pKa = 6.68DD139 pKa = 3.91MGQSVTPLFKK149 pKa = 10.63SVPEE153 pKa = 4.15SVQKK157 pKa = 10.67DD158 pKa = 3.43DD159 pKa = 5.0RR160 pKa = 11.84LYY162 pKa = 10.98EE163 pKa = 4.04YY164 pKa = 10.78LALADD169 pKa = 5.51AIRR172 pKa = 11.84LGNQRR177 pKa = 11.84EE178 pKa = 4.1AGLAAEE184 pKa = 4.69RR185 pKa = 11.84LSEE188 pKa = 3.9GLRR191 pKa = 11.84RR192 pKa = 11.84NDD194 pKa = 3.15QHH196 pKa = 6.6PRR198 pKa = 11.84PAACNAKK205 pKa = 9.61GCRR208 pKa = 11.84GSAGRR213 pKa = 11.84HH214 pKa = 3.92LRR216 pKa = 11.84NRR218 pKa = 11.84LVFVGGCTTALFITDD233 pKa = 3.88SVTLEE238 pKa = 4.06NVRR241 pKa = 11.84ATDD244 pKa = 3.98DD245 pKa = 3.14VDD247 pKa = 6.39LIVDD251 pKa = 4.3LAGFSEE257 pKa = 4.69WAQLPGGGLALGAARR272 pKa = 11.84GGGRR276 pKa = 11.84RR277 pKa = 11.84QQDD280 pKa = 3.15RR281 pKa = 11.84ARR283 pKa = 11.84RR284 pKa = 11.84RR285 pKa = 11.84CRR287 pKa = 11.84FVAAGIAARR296 pKa = 11.84TSGKK300 pKa = 9.25VLWCVTQQDD309 pKa = 3.82LFAPVAHH316 pKa = 6.18QAGLPPSRR324 pKa = 11.84VIHH327 pKa = 5.88VEE329 pKa = 3.63AGDD332 pKa = 3.78EE333 pKa = 4.22KK334 pKa = 10.37TVLGMEE340 pKa = 4.91EE341 pKa = 4.39GRR343 pKa = 11.84QSARR347 pKa = 11.84ARR349 pKa = 11.84PVYY352 pKa = 10.35NSRR355 pKa = 3.6
MM1 pKa = 7.67KK2 pKa = 10.47SQDD5 pKa = 3.45HH6 pKa = 6.27RR7 pKa = 11.84HH8 pKa = 5.15TAQARR13 pKa = 11.84QPTRR17 pKa = 11.84AGEE20 pKa = 4.18RR21 pKa = 11.84KK22 pKa = 9.15GCRR25 pKa = 11.84SSALGSPGRR34 pKa = 11.84RR35 pKa = 11.84SVEE38 pKa = 3.64DD39 pKa = 3.98PYY41 pKa = 11.44SVRR44 pKa = 11.84NLGSSLGISKK54 pKa = 9.06TEE56 pKa = 3.8VNASINRR63 pKa = 11.84SLASGIAARR72 pKa = 11.84DD73 pKa = 3.59RR74 pKa = 11.84DD75 pKa = 3.7TGRR78 pKa = 11.84AKK80 pKa = 10.44PSRR83 pKa = 11.84RR84 pKa = 11.84SLLNFIVHH92 pKa = 6.0GLKK95 pKa = 10.31FVFPAKK101 pKa = 9.87PGAMQRR107 pKa = 11.84GIPTAFAAPVLKK119 pKa = 10.61NALISAGSYY128 pKa = 9.73IYY130 pKa = 10.25VWPYY134 pKa = 11.59ASGHH138 pKa = 6.68DD139 pKa = 3.91MGQSVTPLFKK149 pKa = 10.63SVPEE153 pKa = 4.15SVQKK157 pKa = 10.67DD158 pKa = 3.43DD159 pKa = 5.0RR160 pKa = 11.84LYY162 pKa = 10.98EE163 pKa = 4.04YY164 pKa = 10.78LALADD169 pKa = 5.51AIRR172 pKa = 11.84LGNQRR177 pKa = 11.84EE178 pKa = 4.1AGLAAEE184 pKa = 4.69RR185 pKa = 11.84LSEE188 pKa = 3.9GLRR191 pKa = 11.84RR192 pKa = 11.84NDD194 pKa = 3.15QHH196 pKa = 6.6PRR198 pKa = 11.84PAACNAKK205 pKa = 9.61GCRR208 pKa = 11.84GSAGRR213 pKa = 11.84HH214 pKa = 3.92LRR216 pKa = 11.84NRR218 pKa = 11.84LVFVGGCTTALFITDD233 pKa = 3.88SVTLEE238 pKa = 4.06NVRR241 pKa = 11.84ATDD244 pKa = 3.98DD245 pKa = 3.14VDD247 pKa = 6.39LIVDD251 pKa = 4.3LAGFSEE257 pKa = 4.69WAQLPGGGLALGAARR272 pKa = 11.84GGGRR276 pKa = 11.84RR277 pKa = 11.84QQDD280 pKa = 3.15RR281 pKa = 11.84ARR283 pKa = 11.84RR284 pKa = 11.84RR285 pKa = 11.84CRR287 pKa = 11.84FVAAGIAARR296 pKa = 11.84TSGKK300 pKa = 9.25VLWCVTQQDD309 pKa = 3.82LFAPVAHH316 pKa = 6.18QAGLPPSRR324 pKa = 11.84VIHH327 pKa = 5.88VEE329 pKa = 3.63AGDD332 pKa = 3.78EE333 pKa = 4.22KK334 pKa = 10.37TVLGMEE340 pKa = 4.91EE341 pKa = 4.39GRR343 pKa = 11.84QSARR347 pKa = 11.84ARR349 pKa = 11.84PVYY352 pKa = 10.35NSRR355 pKa = 3.6
Molecular weight: 38.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1794598 |
39 |
2854 |
293.6 |
31.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.286 ± 0.045 | 0.819 ± 0.01 |
5.687 ± 0.024 | 5.809 ± 0.029 |
3.909 ± 0.022 | 8.491 ± 0.036 |
2.012 ± 0.016 | 5.371 ± 0.024 |
3.723 ± 0.023 | 9.797 ± 0.031 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.516 ± 0.015 | 2.752 ± 0.019 |
5.062 ± 0.025 | 3.006 ± 0.017 |
6.998 ± 0.035 | 5.514 ± 0.019 |
5.223 ± 0.024 | 7.428 ± 0.025 |
1.347 ± 0.013 | 2.251 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
