
Arthrobacter sp. PAMC 25486
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Micrococcaceae; Arthrobacter; unclassified Arthrobacter
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
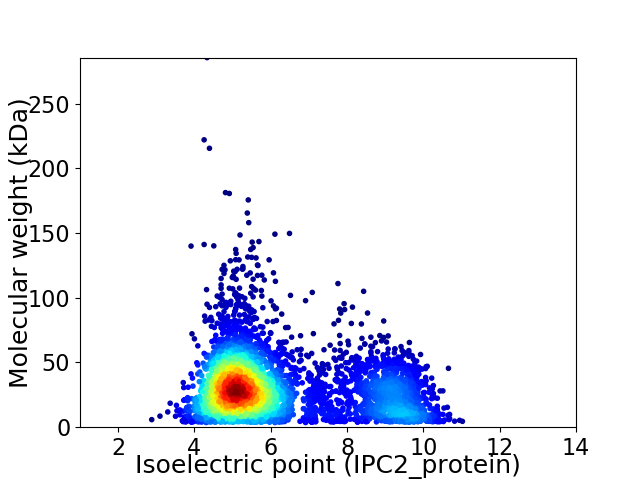
Virtual 2D-PAGE plot for 4345 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A1D2R9|A0A0A1D2R9_9MICC Cytochrome c biogenesis protein CcdA OS=Arthrobacter sp. PAMC 25486 OX=1494608 GN=ART_3115 PE=4 SV=1
MM1 pKa = 7.72GEE3 pKa = 4.48HH4 pKa = 7.8PIQSSDD10 pKa = 2.98NSAAPEE16 pKa = 4.6GGQQPGNQPQPEE28 pKa = 4.19NQQPEE33 pKa = 4.11QFAAQPGGLPPPFAQPPFYY52 pKa = 9.86PPPFAQPGDD61 pKa = 3.65QQGPYY66 pKa = 8.38PQSPHH71 pKa = 5.41SQPYY75 pKa = 8.39PPYY78 pKa = 9.91GAPALKK84 pKa = 10.06QDD86 pKa = 3.98TTNTMAIVAFVLSFVVNLAGVITGHH111 pKa = 6.85IALGQIKK118 pKa = 8.36RR119 pKa = 11.84TGEE122 pKa = 3.82KK123 pKa = 10.62GRR125 pKa = 11.84GFALAGVIIGYY136 pKa = 9.96VSLAASVAAAIILVVVLAAGGLAASGVTSYY166 pKa = 11.73DD167 pKa = 3.23PDD169 pKa = 4.72AEE171 pKa = 4.28SARR174 pKa = 11.84PNGTVEE180 pKa = 4.06QVNTNAYY187 pKa = 10.19GGITFGRR194 pKa = 11.84DD195 pKa = 2.9GVLIPPTVLTADD207 pKa = 3.24VDD209 pKa = 3.27IWDD212 pKa = 4.52LPKK215 pKa = 10.87EE216 pKa = 4.15PASLADD222 pKa = 3.38IGIKK226 pKa = 10.32ASASGEE232 pKa = 4.0PVQVVVYY239 pKa = 10.29LDD241 pKa = 4.78FMCPDD246 pKa = 3.51CADD249 pKa = 3.58FEE251 pKa = 4.65EE252 pKa = 5.54EE253 pKa = 4.25YY254 pKa = 11.14GPSLRR259 pKa = 11.84ALGDD263 pKa = 3.33QGGVTLEE270 pKa = 3.8YY271 pKa = 10.62RR272 pKa = 11.84PGGFLDD278 pKa = 4.54EE279 pKa = 4.67YY280 pKa = 11.26SDD282 pKa = 3.91GADD285 pKa = 3.37YY286 pKa = 11.05SSRR289 pKa = 11.84AAEE292 pKa = 4.06AAACVANTAPEE303 pKa = 4.33DD304 pKa = 3.57YY305 pKa = 10.65KK306 pKa = 11.29RR307 pKa = 11.84FIDD310 pKa = 3.85TLLANQPAIDD320 pKa = 4.69SMMPDD325 pKa = 2.97TNYY328 pKa = 10.73LVEE331 pKa = 4.21LANAVGAGDD340 pKa = 5.22IDD342 pKa = 4.31DD343 pKa = 5.3CLAGMDD349 pKa = 4.1MFDD352 pKa = 4.09LVSGATLLAVGHH364 pKa = 6.65GLTGIPGVFMDD375 pKa = 4.77GKK377 pKa = 10.38QWMDD381 pKa = 3.37GPFPDD386 pKa = 5.03FAQGVLDD393 pKa = 4.63AKK395 pKa = 10.61QQ396 pKa = 2.8
MM1 pKa = 7.72GEE3 pKa = 4.48HH4 pKa = 7.8PIQSSDD10 pKa = 2.98NSAAPEE16 pKa = 4.6GGQQPGNQPQPEE28 pKa = 4.19NQQPEE33 pKa = 4.11QFAAQPGGLPPPFAQPPFYY52 pKa = 9.86PPPFAQPGDD61 pKa = 3.65QQGPYY66 pKa = 8.38PQSPHH71 pKa = 5.41SQPYY75 pKa = 8.39PPYY78 pKa = 9.91GAPALKK84 pKa = 10.06QDD86 pKa = 3.98TTNTMAIVAFVLSFVVNLAGVITGHH111 pKa = 6.85IALGQIKK118 pKa = 8.36RR119 pKa = 11.84TGEE122 pKa = 3.82KK123 pKa = 10.62GRR125 pKa = 11.84GFALAGVIIGYY136 pKa = 9.96VSLAASVAAAIILVVVLAAGGLAASGVTSYY166 pKa = 11.73DD167 pKa = 3.23PDD169 pKa = 4.72AEE171 pKa = 4.28SARR174 pKa = 11.84PNGTVEE180 pKa = 4.06QVNTNAYY187 pKa = 10.19GGITFGRR194 pKa = 11.84DD195 pKa = 2.9GVLIPPTVLTADD207 pKa = 3.24VDD209 pKa = 3.27IWDD212 pKa = 4.52LPKK215 pKa = 10.87EE216 pKa = 4.15PASLADD222 pKa = 3.38IGIKK226 pKa = 10.32ASASGEE232 pKa = 4.0PVQVVVYY239 pKa = 10.29LDD241 pKa = 4.78FMCPDD246 pKa = 3.51CADD249 pKa = 3.58FEE251 pKa = 4.65EE252 pKa = 5.54EE253 pKa = 4.25YY254 pKa = 11.14GPSLRR259 pKa = 11.84ALGDD263 pKa = 3.33QGGVTLEE270 pKa = 3.8YY271 pKa = 10.62RR272 pKa = 11.84PGGFLDD278 pKa = 4.54EE279 pKa = 4.67YY280 pKa = 11.26SDD282 pKa = 3.91GADD285 pKa = 3.37YY286 pKa = 11.05SSRR289 pKa = 11.84AAEE292 pKa = 4.06AAACVANTAPEE303 pKa = 4.33DD304 pKa = 3.57YY305 pKa = 10.65KK306 pKa = 11.29RR307 pKa = 11.84FIDD310 pKa = 3.85TLLANQPAIDD320 pKa = 4.69SMMPDD325 pKa = 2.97TNYY328 pKa = 10.73LVEE331 pKa = 4.21LANAVGAGDD340 pKa = 5.22IDD342 pKa = 4.31DD343 pKa = 5.3CLAGMDD349 pKa = 4.1MFDD352 pKa = 4.09LVSGATLLAVGHH364 pKa = 6.65GLTGIPGVFMDD375 pKa = 4.77GKK377 pKa = 10.38QWMDD381 pKa = 3.37GPFPDD386 pKa = 5.03FAQGVLDD393 pKa = 4.63AKK395 pKa = 10.61QQ396 pKa = 2.8
Molecular weight: 41.17 kDa
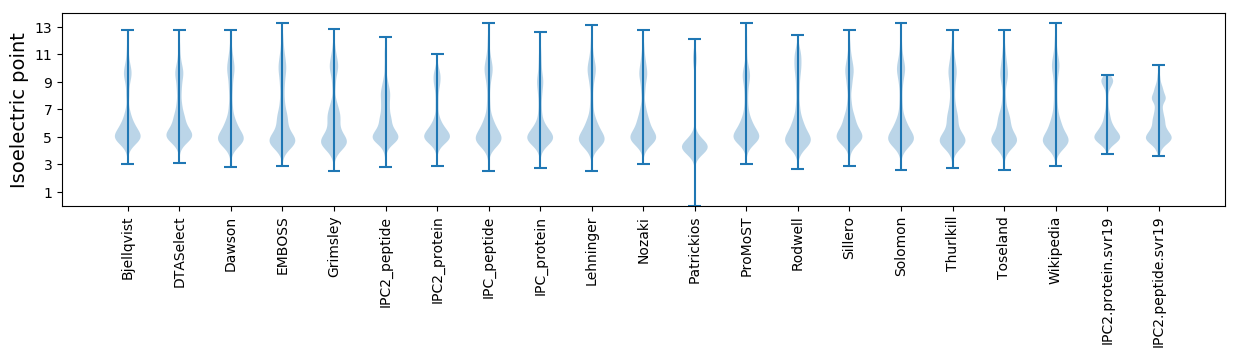
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A1CRI7|A0A0A1CRI7_9MICC Arylsulfatase OS=Arthrobacter sp. PAMC 25486 OX=1494608 GN=ART_0141 PE=4 SV=1
MM1 pKa = 7.59ALGRR5 pKa = 11.84QVPPSFLHH13 pKa = 5.95ARR15 pKa = 11.84KK16 pKa = 7.42TVLGRR21 pKa = 11.84ILRR24 pKa = 11.84LHH26 pKa = 7.07PAQKK30 pKa = 10.18LVARR34 pKa = 11.84RR35 pKa = 11.84FTRR38 pKa = 11.84QQ39 pKa = 2.67
MM1 pKa = 7.59ALGRR5 pKa = 11.84QVPPSFLHH13 pKa = 5.95ARR15 pKa = 11.84KK16 pKa = 7.42TVLGRR21 pKa = 11.84ILRR24 pKa = 11.84LHH26 pKa = 7.07PAQKK30 pKa = 10.18LVARR34 pKa = 11.84RR35 pKa = 11.84FTRR38 pKa = 11.84QQ39 pKa = 2.67
Molecular weight: 4.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1309562 |
37 |
2773 |
301.4 |
32.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.074 ± 0.053 | 0.623 ± 0.009 |
5.434 ± 0.027 | 5.449 ± 0.03 |
3.31 ± 0.021 | 8.947 ± 0.034 |
2.156 ± 0.017 | 4.583 ± 0.029 |
2.918 ± 0.027 | 10.253 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.276 ± 0.017 | 2.699 ± 0.022 |
5.357 ± 0.027 | 3.173 ± 0.023 |
5.879 ± 0.036 | 6.017 ± 0.026 |
6.058 ± 0.033 | 8.28 ± 0.032 |
1.469 ± 0.017 | 2.044 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
