
Oceanobacillus halophilus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Oceanobacillus
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
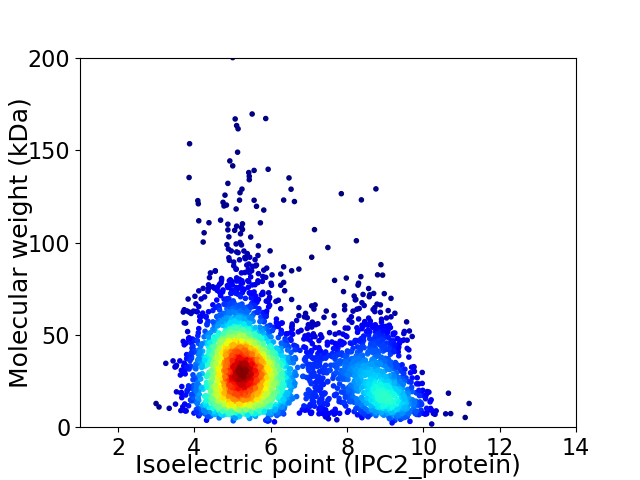
Virtual 2D-PAGE plot for 3727 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A495AB14|A0A495AB14_9BACI Chromate transporter OS=Oceanobacillus halophilus OX=930130 GN=D8M06_05090 PE=3 SV=1
MM1 pKa = 6.96MVIAFVLAACSGSNTSDD18 pKa = 2.72TGEE21 pKa = 3.83AVQEE25 pKa = 4.37GNNPTEE31 pKa = 4.47SANSQPGTEE40 pKa = 3.98NVLRR44 pKa = 11.84IAMSTEE50 pKa = 3.57VDD52 pKa = 3.41NLDD55 pKa = 4.18PYY57 pKa = 10.93ISAATDD63 pKa = 3.53TQSMMDD69 pKa = 3.54NVFDD73 pKa = 4.43GLLDD77 pKa = 3.62TDD79 pKa = 3.91EE80 pKa = 4.64SGKK83 pKa = 10.24LVPAIAEE90 pKa = 4.28DD91 pKa = 4.07YY92 pKa = 10.18EE93 pKa = 4.32ISEE96 pKa = 5.33DD97 pKa = 3.41GLTYY101 pKa = 10.16TFQLKK106 pKa = 10.31EE107 pKa = 4.44GILFHH112 pKa = 7.44DD113 pKa = 4.84GSEE116 pKa = 4.51LISEE120 pKa = 4.46DD121 pKa = 3.06VVYY124 pKa = 10.53SYY126 pKa = 11.61EE127 pKa = 4.11RR128 pKa = 11.84LSGLNGEE135 pKa = 5.0EE136 pKa = 4.13PLSSQFAGIEE146 pKa = 4.13NIEE149 pKa = 4.23APSDD153 pKa = 3.85YY154 pKa = 10.95EE155 pKa = 4.17VVITLKK161 pKa = 10.59EE162 pKa = 3.97NNSAFLAANIRR173 pKa = 11.84PILPAGYY180 pKa = 9.16EE181 pKa = 4.1EE182 pKa = 5.11QSTSPIGAGPFKK194 pKa = 10.78FEE196 pKa = 4.61EE197 pKa = 4.42YY198 pKa = 10.19QAGQQLTLVKK208 pKa = 10.95NEE210 pKa = 4.4DD211 pKa = 3.99YY212 pKa = 11.58YY213 pKa = 11.18MQDD216 pKa = 3.03KK217 pKa = 10.62VPPFDD222 pKa = 3.29KK223 pKa = 10.93VQFIVMPDD231 pKa = 3.45PEE233 pKa = 4.56SAVLAMQAGEE243 pKa = 4.07IDD245 pKa = 4.68VIPGVTPQGMMQLGEE260 pKa = 4.35SVNTVSGPQNMVQLMALNNEE280 pKa = 4.26VEE282 pKa = 4.36PLDD285 pKa = 4.01DD286 pKa = 3.93VNVRR290 pKa = 11.84KK291 pKa = 10.03AINLAIDD298 pKa = 3.38KK299 pKa = 11.0DD300 pKa = 4.12MIIEE304 pKa = 4.31TVADD308 pKa = 3.75GMGTKK313 pKa = 10.32LGSNFSPAMDD323 pKa = 4.52FYY325 pKa = 11.75YY326 pKa = 10.56EE327 pKa = 4.72AGLEE331 pKa = 4.7DD332 pKa = 4.59YY333 pKa = 8.99YY334 pKa = 11.37QPNIEE339 pKa = 4.09EE340 pKa = 4.36AKK342 pKa = 10.86ALLADD347 pKa = 4.25AGYY350 pKa = 11.43ADD352 pKa = 4.97GFDD355 pKa = 4.69LEE357 pKa = 4.66LTVPSDD363 pKa = 3.67YY364 pKa = 10.78QFHH367 pKa = 6.93VDD369 pKa = 3.22TAQVVVEE376 pKa = 4.02QLSQIGINVEE386 pKa = 3.49IKK388 pKa = 10.67LIEE391 pKa = 4.23FSSWLEE397 pKa = 3.7TVYY400 pKa = 11.01QDD402 pKa = 3.46AQYY405 pKa = 10.95EE406 pKa = 4.39STIVSFTGKK415 pKa = 10.04LDD417 pKa = 3.77PYY419 pKa = 9.44EE420 pKa = 3.94VLVRR424 pKa = 11.84YY425 pKa = 10.09ISDD428 pKa = 3.62YY429 pKa = 10.73QSNFVNYY436 pKa = 10.21NNPEE440 pKa = 3.68YY441 pKa = 10.69DD442 pKa = 3.52ALIEE446 pKa = 4.01QAVASTDD453 pKa = 3.62EE454 pKa = 4.61NEE456 pKa = 4.07MADD459 pKa = 4.58LYY461 pKa = 11.19KK462 pKa = 10.28QAQRR466 pKa = 11.84LLTEE470 pKa = 4.81DD471 pKa = 3.45AASVYY476 pKa = 10.7IMDD479 pKa = 5.14PDD481 pKa = 2.98RR482 pKa = 11.84TYY484 pKa = 11.77ALRR487 pKa = 11.84SGLEE491 pKa = 3.75GWKK494 pKa = 9.27MYY496 pKa = 9.85PIQKK500 pKa = 10.05FNLEE504 pKa = 4.58DD505 pKa = 3.41IRR507 pKa = 11.84VTEE510 pKa = 4.06
MM1 pKa = 6.96MVIAFVLAACSGSNTSDD18 pKa = 2.72TGEE21 pKa = 3.83AVQEE25 pKa = 4.37GNNPTEE31 pKa = 4.47SANSQPGTEE40 pKa = 3.98NVLRR44 pKa = 11.84IAMSTEE50 pKa = 3.57VDD52 pKa = 3.41NLDD55 pKa = 4.18PYY57 pKa = 10.93ISAATDD63 pKa = 3.53TQSMMDD69 pKa = 3.54NVFDD73 pKa = 4.43GLLDD77 pKa = 3.62TDD79 pKa = 3.91EE80 pKa = 4.64SGKK83 pKa = 10.24LVPAIAEE90 pKa = 4.28DD91 pKa = 4.07YY92 pKa = 10.18EE93 pKa = 4.32ISEE96 pKa = 5.33DD97 pKa = 3.41GLTYY101 pKa = 10.16TFQLKK106 pKa = 10.31EE107 pKa = 4.44GILFHH112 pKa = 7.44DD113 pKa = 4.84GSEE116 pKa = 4.51LISEE120 pKa = 4.46DD121 pKa = 3.06VVYY124 pKa = 10.53SYY126 pKa = 11.61EE127 pKa = 4.11RR128 pKa = 11.84LSGLNGEE135 pKa = 5.0EE136 pKa = 4.13PLSSQFAGIEE146 pKa = 4.13NIEE149 pKa = 4.23APSDD153 pKa = 3.85YY154 pKa = 10.95EE155 pKa = 4.17VVITLKK161 pKa = 10.59EE162 pKa = 3.97NNSAFLAANIRR173 pKa = 11.84PILPAGYY180 pKa = 9.16EE181 pKa = 4.1EE182 pKa = 5.11QSTSPIGAGPFKK194 pKa = 10.78FEE196 pKa = 4.61EE197 pKa = 4.42YY198 pKa = 10.19QAGQQLTLVKK208 pKa = 10.95NEE210 pKa = 4.4DD211 pKa = 3.99YY212 pKa = 11.58YY213 pKa = 11.18MQDD216 pKa = 3.03KK217 pKa = 10.62VPPFDD222 pKa = 3.29KK223 pKa = 10.93VQFIVMPDD231 pKa = 3.45PEE233 pKa = 4.56SAVLAMQAGEE243 pKa = 4.07IDD245 pKa = 4.68VIPGVTPQGMMQLGEE260 pKa = 4.35SVNTVSGPQNMVQLMALNNEE280 pKa = 4.26VEE282 pKa = 4.36PLDD285 pKa = 4.01DD286 pKa = 3.93VNVRR290 pKa = 11.84KK291 pKa = 10.03AINLAIDD298 pKa = 3.38KK299 pKa = 11.0DD300 pKa = 4.12MIIEE304 pKa = 4.31TVADD308 pKa = 3.75GMGTKK313 pKa = 10.32LGSNFSPAMDD323 pKa = 4.52FYY325 pKa = 11.75YY326 pKa = 10.56EE327 pKa = 4.72AGLEE331 pKa = 4.7DD332 pKa = 4.59YY333 pKa = 8.99YY334 pKa = 11.37QPNIEE339 pKa = 4.09EE340 pKa = 4.36AKK342 pKa = 10.86ALLADD347 pKa = 4.25AGYY350 pKa = 11.43ADD352 pKa = 4.97GFDD355 pKa = 4.69LEE357 pKa = 4.66LTVPSDD363 pKa = 3.67YY364 pKa = 10.78QFHH367 pKa = 6.93VDD369 pKa = 3.22TAQVVVEE376 pKa = 4.02QLSQIGINVEE386 pKa = 3.49IKK388 pKa = 10.67LIEE391 pKa = 4.23FSSWLEE397 pKa = 3.7TVYY400 pKa = 11.01QDD402 pKa = 3.46AQYY405 pKa = 10.95EE406 pKa = 4.39STIVSFTGKK415 pKa = 10.04LDD417 pKa = 3.77PYY419 pKa = 9.44EE420 pKa = 3.94VLVRR424 pKa = 11.84YY425 pKa = 10.09ISDD428 pKa = 3.62YY429 pKa = 10.73QSNFVNYY436 pKa = 10.21NNPEE440 pKa = 3.68YY441 pKa = 10.69DD442 pKa = 3.52ALIEE446 pKa = 4.01QAVASTDD453 pKa = 3.62EE454 pKa = 4.61NEE456 pKa = 4.07MADD459 pKa = 4.58LYY461 pKa = 11.19KK462 pKa = 10.28QAQRR466 pKa = 11.84LLTEE470 pKa = 4.81DD471 pKa = 3.45AASVYY476 pKa = 10.7IMDD479 pKa = 5.14PDD481 pKa = 2.98RR482 pKa = 11.84TYY484 pKa = 11.77ALRR487 pKa = 11.84SGLEE491 pKa = 3.75GWKK494 pKa = 9.27MYY496 pKa = 9.85PIQKK500 pKa = 10.05FNLEE504 pKa = 4.58DD505 pKa = 3.41IRR507 pKa = 11.84VTEE510 pKa = 4.06
Molecular weight: 56.43 kDa
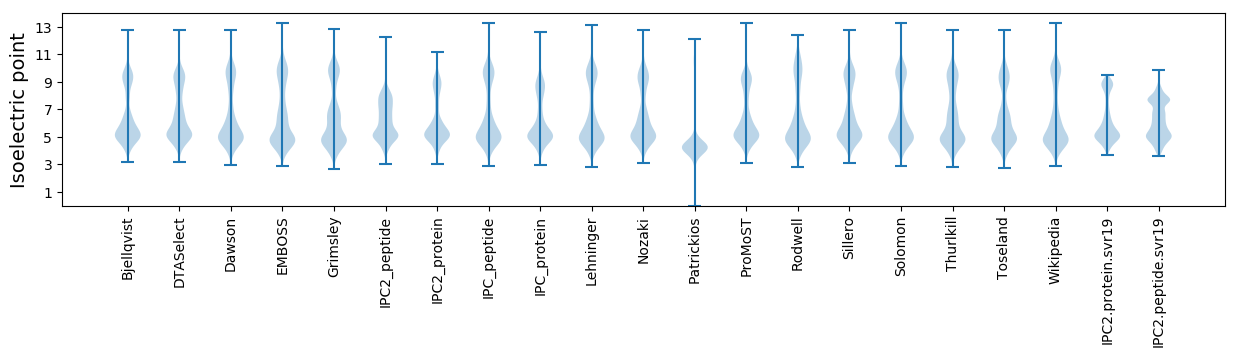
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A494ZVK6|A0A494ZVK6_9BACI Bacteriocin aureocin A53 OS=Oceanobacillus halophilus OX=930130 GN=D8M06_16260 PE=4 SV=1
MM1 pKa = 7.26NRR3 pKa = 11.84TNPSINRR10 pKa = 11.84TNPSINRR17 pKa = 11.84TNPSINRR24 pKa = 11.84TNPSINRR31 pKa = 11.84TNPSINRR38 pKa = 11.84TNPSINRR45 pKa = 11.84TNPSINRR52 pKa = 11.84TNPSINRR59 pKa = 11.84TNPSINRR66 pKa = 11.84TSPSINRR73 pKa = 11.84TSPSINRR80 pKa = 11.84TNPSINHH87 pKa = 6.18TSPSINRR94 pKa = 11.84LQASTHH100 pKa = 5.41KK101 pKa = 10.8QPFAPFYY108 pKa = 10.25SKK110 pKa = 11.22QNIRR114 pKa = 3.69
MM1 pKa = 7.26NRR3 pKa = 11.84TNPSINRR10 pKa = 11.84TNPSINRR17 pKa = 11.84TNPSINRR24 pKa = 11.84TNPSINRR31 pKa = 11.84TNPSINRR38 pKa = 11.84TNPSINRR45 pKa = 11.84TNPSINRR52 pKa = 11.84TNPSINRR59 pKa = 11.84TNPSINRR66 pKa = 11.84TSPSINRR73 pKa = 11.84TSPSINRR80 pKa = 11.84TNPSINHH87 pKa = 6.18TSPSINRR94 pKa = 11.84LQASTHH100 pKa = 5.41KK101 pKa = 10.8QPFAPFYY108 pKa = 10.25SKK110 pKa = 11.22QNIRR114 pKa = 3.69
Molecular weight: 12.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1103683 |
14 |
1747 |
296.1 |
33.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.563 ± 0.04 | 0.591 ± 0.011 |
5.325 ± 0.04 | 7.813 ± 0.055 |
4.525 ± 0.039 | 6.772 ± 0.037 |
2.012 ± 0.019 | 8.271 ± 0.048 |
6.894 ± 0.039 | 9.5 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.831 ± 0.02 | 4.813 ± 0.024 |
3.516 ± 0.024 | 3.625 ± 0.024 |
3.868 ± 0.03 | 6.02 ± 0.029 |
5.475 ± 0.026 | 6.97 ± 0.03 |
0.991 ± 0.014 | 3.624 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
