
Parvularcula bermudensis (strain ATCC BAA-594 / HTCC2503 / KCTC 12087)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Parvularculales; Parvularculaceae; Parvularcula; Parvularcula bermudensis
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins
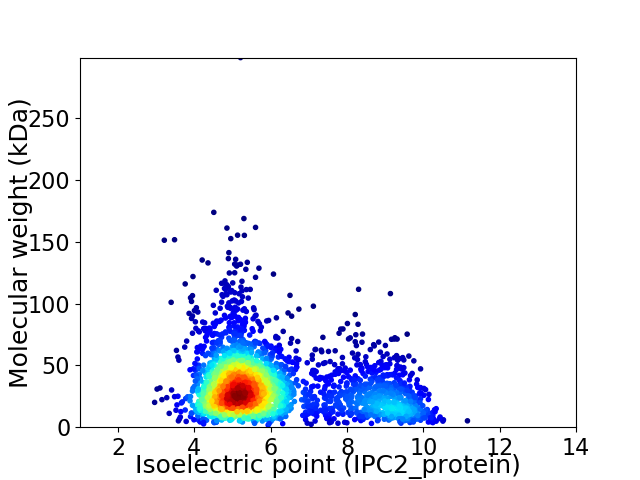
Virtual 2D-PAGE plot for 2687 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E0TEQ1|E0TEQ1_PARBH Transcription termination/antitermination protein NusA OS=Parvularcula bermudensis (strain ATCC BAA-594 / HTCC2503 / KCTC 12087) OX=314260 GN=nusA PE=3 SV=1
MM1 pKa = 7.68RR2 pKa = 11.84LSTGLFAASSLIALAACSSTGGTPDD27 pKa = 5.33PILGEE32 pKa = 4.76DD33 pKa = 4.12GQPAPPRR40 pKa = 11.84NLVTNALSYY49 pKa = 10.28RR50 pKa = 11.84AEE52 pKa = 4.03NHH54 pKa = 5.62GSAVSEE60 pKa = 4.42TYY62 pKa = 10.97DD63 pKa = 3.7LAVTAMRR70 pKa = 11.84TRR72 pKa = 11.84YY73 pKa = 6.24QTNGAAVFVGIEE85 pKa = 3.8HH86 pKa = 7.07DD87 pKa = 3.69AVGEE91 pKa = 4.28SEE93 pKa = 4.36TTEE96 pKa = 3.87FRR98 pKa = 11.84YY99 pKa = 10.3DD100 pKa = 3.09AAADD104 pKa = 3.78TFTIDD109 pKa = 4.23IDD111 pKa = 4.74LPNADD116 pKa = 4.98DD117 pKa = 5.58APILVNEE124 pKa = 4.23TWGPLLLLTPKK135 pKa = 10.75DD136 pKa = 3.74LFGLNNGVSAVYY148 pKa = 9.55LAKK151 pKa = 10.68QPDD154 pKa = 4.1FYY156 pKa = 11.73GFVDD160 pKa = 3.56EE161 pKa = 4.88TLIGGNEE168 pKa = 4.0FTADD172 pKa = 3.52AYY174 pKa = 10.08IDD176 pKa = 3.55ALGAVDD182 pKa = 3.73ATGSGSGYY190 pKa = 9.52LALWVTIANDD200 pKa = 3.56LGFEE204 pKa = 4.08GSEE207 pKa = 3.9RR208 pKa = 11.84DD209 pKa = 3.84VGSLPIYY216 pKa = 9.64IANDD220 pKa = 3.49PATFGAPGSLAGDD233 pKa = 3.82IEE235 pKa = 4.25AATAYY240 pKa = 10.91VEE242 pKa = 4.44DD243 pKa = 4.98LSPAAFSDD251 pKa = 3.72AQAAAPVPSSAFDD264 pKa = 4.54PIMSEE269 pKa = 4.4LLATAPEE276 pKa = 4.32SFGVNVEE283 pKa = 4.18GQISDD288 pKa = 3.56NDD290 pKa = 3.37ASFIIAVMDD299 pKa = 4.17DD300 pKa = 3.45VGGTAVEE307 pKa = 4.68DD308 pKa = 4.38YY309 pKa = 9.36ITAITIITDD318 pKa = 4.36DD319 pKa = 3.92IEE321 pKa = 4.2AAPNYY326 pKa = 9.83LYY328 pKa = 10.36QANGVTYY335 pKa = 9.48YY336 pKa = 10.42QYY338 pKa = 9.66KK339 pKa = 9.07TEE341 pKa = 4.14GDD343 pKa = 3.82VVDD346 pKa = 4.1TRR348 pKa = 11.84FVAAGEE354 pKa = 4.18WQKK357 pKa = 9.76TEE359 pKa = 3.91ASGEE363 pKa = 4.33TIFGHH368 pKa = 6.0FVYY371 pKa = 10.18GQLTDD376 pKa = 4.02PDD378 pKa = 4.05EE379 pKa = 4.64MPTSGTASYY388 pKa = 9.61TGTVYY393 pKa = 11.04GSILRR398 pKa = 11.84QNRR401 pKa = 11.84VDD403 pKa = 3.62SLRR406 pKa = 11.84GGLNLNTDD414 pKa = 4.8FQTGALDD421 pKa = 3.38MTFNADD427 pKa = 2.46IAYY430 pKa = 9.98RR431 pKa = 11.84DD432 pKa = 4.22DD433 pKa = 3.76EE434 pKa = 5.57GITQFIDD441 pKa = 3.43YY442 pKa = 10.96ADD444 pKa = 3.6FTGTGRR450 pKa = 11.84IDD452 pKa = 3.59DD453 pKa = 4.45ANFDD457 pKa = 3.63GTMQGVTSIDD467 pKa = 3.29TSEE470 pKa = 4.06TDD472 pKa = 2.97SPLVVDD478 pKa = 5.73GSFEE482 pKa = 3.98GHH484 pKa = 6.52FYY486 pKa = 11.28GPTAQEE492 pKa = 3.71AGGTFEE498 pKa = 5.12FSNTEE503 pKa = 3.56AAATGSFVAVRR514 pKa = 11.84DD515 pKa = 3.73
MM1 pKa = 7.68RR2 pKa = 11.84LSTGLFAASSLIALAACSSTGGTPDD27 pKa = 5.33PILGEE32 pKa = 4.76DD33 pKa = 4.12GQPAPPRR40 pKa = 11.84NLVTNALSYY49 pKa = 10.28RR50 pKa = 11.84AEE52 pKa = 4.03NHH54 pKa = 5.62GSAVSEE60 pKa = 4.42TYY62 pKa = 10.97DD63 pKa = 3.7LAVTAMRR70 pKa = 11.84TRR72 pKa = 11.84YY73 pKa = 6.24QTNGAAVFVGIEE85 pKa = 3.8HH86 pKa = 7.07DD87 pKa = 3.69AVGEE91 pKa = 4.28SEE93 pKa = 4.36TTEE96 pKa = 3.87FRR98 pKa = 11.84YY99 pKa = 10.3DD100 pKa = 3.09AAADD104 pKa = 3.78TFTIDD109 pKa = 4.23IDD111 pKa = 4.74LPNADD116 pKa = 4.98DD117 pKa = 5.58APILVNEE124 pKa = 4.23TWGPLLLLTPKK135 pKa = 10.75DD136 pKa = 3.74LFGLNNGVSAVYY148 pKa = 9.55LAKK151 pKa = 10.68QPDD154 pKa = 4.1FYY156 pKa = 11.73GFVDD160 pKa = 3.56EE161 pKa = 4.88TLIGGNEE168 pKa = 4.0FTADD172 pKa = 3.52AYY174 pKa = 10.08IDD176 pKa = 3.55ALGAVDD182 pKa = 3.73ATGSGSGYY190 pKa = 9.52LALWVTIANDD200 pKa = 3.56LGFEE204 pKa = 4.08GSEE207 pKa = 3.9RR208 pKa = 11.84DD209 pKa = 3.84VGSLPIYY216 pKa = 9.64IANDD220 pKa = 3.49PATFGAPGSLAGDD233 pKa = 3.82IEE235 pKa = 4.25AATAYY240 pKa = 10.91VEE242 pKa = 4.44DD243 pKa = 4.98LSPAAFSDD251 pKa = 3.72AQAAAPVPSSAFDD264 pKa = 4.54PIMSEE269 pKa = 4.4LLATAPEE276 pKa = 4.32SFGVNVEE283 pKa = 4.18GQISDD288 pKa = 3.56NDD290 pKa = 3.37ASFIIAVMDD299 pKa = 4.17DD300 pKa = 3.45VGGTAVEE307 pKa = 4.68DD308 pKa = 4.38YY309 pKa = 9.36ITAITIITDD318 pKa = 4.36DD319 pKa = 3.92IEE321 pKa = 4.2AAPNYY326 pKa = 9.83LYY328 pKa = 10.36QANGVTYY335 pKa = 9.48YY336 pKa = 10.42QYY338 pKa = 9.66KK339 pKa = 9.07TEE341 pKa = 4.14GDD343 pKa = 3.82VVDD346 pKa = 4.1TRR348 pKa = 11.84FVAAGEE354 pKa = 4.18WQKK357 pKa = 9.76TEE359 pKa = 3.91ASGEE363 pKa = 4.33TIFGHH368 pKa = 6.0FVYY371 pKa = 10.18GQLTDD376 pKa = 4.02PDD378 pKa = 4.05EE379 pKa = 4.64MPTSGTASYY388 pKa = 9.61TGTVYY393 pKa = 11.04GSILRR398 pKa = 11.84QNRR401 pKa = 11.84VDD403 pKa = 3.62SLRR406 pKa = 11.84GGLNLNTDD414 pKa = 4.8FQTGALDD421 pKa = 3.38MTFNADD427 pKa = 2.46IAYY430 pKa = 9.98RR431 pKa = 11.84DD432 pKa = 4.22DD433 pKa = 3.76EE434 pKa = 5.57GITQFIDD441 pKa = 3.43YY442 pKa = 10.96ADD444 pKa = 3.6FTGTGRR450 pKa = 11.84IDD452 pKa = 3.59DD453 pKa = 4.45ANFDD457 pKa = 3.63GTMQGVTSIDD467 pKa = 3.29TSEE470 pKa = 4.06TDD472 pKa = 2.97SPLVVDD478 pKa = 5.73GSFEE482 pKa = 3.98GHH484 pKa = 6.52FYY486 pKa = 11.28GPTAQEE492 pKa = 3.71AGGTFEE498 pKa = 5.12FSNTEE503 pKa = 3.56AAATGSFVAVRR514 pKa = 11.84DD515 pKa = 3.73
Molecular weight: 54.22 kDa
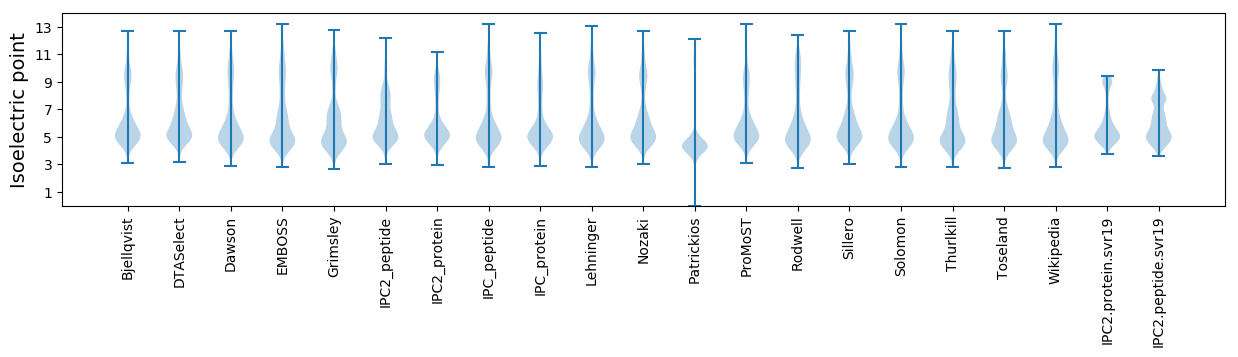
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E0TGT2|E0TGT2_PARBH Anthranilate phosphoribosyltransferase OS=Parvularcula bermudensis (strain ATCC BAA-594 / HTCC2503 / KCTC 12087) OX=314260 GN=trpD PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 10.14QPSVLKK11 pKa = 10.35RR12 pKa = 11.84KK13 pKa = 9.12RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.52GFRR19 pKa = 11.84ARR21 pKa = 11.84KK22 pKa = 7.68ATVGGRR28 pKa = 11.84RR29 pKa = 11.84VLAARR34 pKa = 11.84RR35 pKa = 11.84SKK37 pKa = 10.53GRR39 pKa = 11.84KK40 pKa = 8.77RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 10.14QPSVLKK11 pKa = 10.35RR12 pKa = 11.84KK13 pKa = 9.12RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.52GFRR19 pKa = 11.84ARR21 pKa = 11.84KK22 pKa = 7.68ATVGGRR28 pKa = 11.84RR29 pKa = 11.84VLAARR34 pKa = 11.84RR35 pKa = 11.84SKK37 pKa = 10.53GRR39 pKa = 11.84KK40 pKa = 8.77RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
870527 |
17 |
2831 |
324.0 |
35.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.18 ± 0.064 | 0.759 ± 0.014 |
6.294 ± 0.04 | 6.28 ± 0.04 |
3.74 ± 0.033 | 8.485 ± 0.043 |
1.891 ± 0.024 | 5.124 ± 0.031 |
3.03 ± 0.044 | 10.339 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.208 ± 0.022 | 2.361 ± 0.026 |
5.268 ± 0.033 | 3.136 ± 0.024 |
7.138 ± 0.048 | 5.618 ± 0.03 |
5.641 ± 0.032 | 7.011 ± 0.041 |
1.205 ± 0.019 | 2.292 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
