
Exophiala mesophila (Black yeast)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; Eurotiomycetes; Chaetothyriomycetidae; Chaetothyriales; Herpotrichiellaceae; Exophiala
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
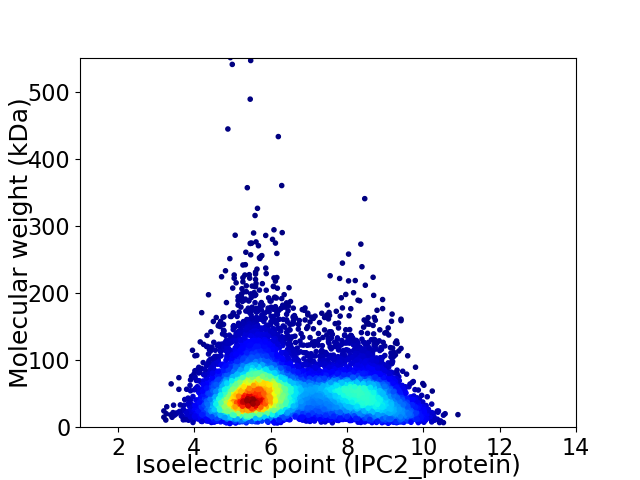
Virtual 2D-PAGE plot for 10000 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D1ZBM4|A0A0D1ZBM4_EXOME Yippee domain-containing protein OS=Exophiala mesophila OX=212818 GN=PV10_06554 PE=3 SV=1
MM1 pKa = 7.61FSTLSISPSMLSISALTLTSLLVPSAAQTFTEE33 pKa = 4.86CNPLTQGGCPPDD45 pKa = 3.94PALGRR50 pKa = 11.84SVSVDD55 pKa = 3.85FTSGPSSEE63 pKa = 4.15FSAVGSTSYY72 pKa = 10.72TSDD75 pKa = 2.85GAAFTVARR83 pKa = 11.84SGDD86 pKa = 3.84SPSITSEE93 pKa = 3.56WYY95 pKa = 9.88IMFGHH100 pKa = 6.54VDD102 pKa = 4.14FVVKK106 pKa = 10.39AAPGTGIVSAAILQSDD122 pKa = 4.62CLDD125 pKa = 4.28EE126 pKa = 6.5IDD128 pKa = 4.05WEE130 pKa = 4.29WLGGDD135 pKa = 3.56NSQAQSNYY143 pKa = 8.23FGKK146 pKa = 11.16GITTTGYY153 pKa = 10.5NRR155 pKa = 11.84GAFHH159 pKa = 6.95GASNNHH165 pKa = 6.09DD166 pKa = 3.5QFHH169 pKa = 6.7TYY171 pKa = 10.38SIDD174 pKa = 2.94WTAEE178 pKa = 3.89QIVWSIDD185 pKa = 3.21GTTVRR190 pKa = 11.84AMSQAQAEE198 pKa = 4.46PGQYY202 pKa = 9.13PQTPMYY208 pKa = 11.08VKK210 pKa = 9.77MGAWAGGDD218 pKa = 3.55PANAPGTIEE227 pKa = 3.88WAGGYY232 pKa = 8.06TDD234 pKa = 4.61YY235 pKa = 11.44SQGPFTMYY243 pKa = 10.22VKK245 pKa = 10.46SVQVTDD251 pKa = 3.63YY252 pKa = 10.25STGSEE257 pKa = 4.08YY258 pKa = 10.36TYY260 pKa = 10.84SGSSGTWQSIQSNGGSINPEE280 pKa = 3.88GKK282 pKa = 8.6GTPVSPDD289 pKa = 2.93SAPAVTSVVPDD300 pKa = 3.41NGAPLGFGNNANRR313 pKa = 11.84EE314 pKa = 4.33SIAATRR320 pKa = 11.84TGYY323 pKa = 9.74PWTGTATASIATASTHH339 pKa = 4.52QTSDD343 pKa = 3.46SSWPSAPASDD353 pKa = 4.56SPSSPIVYY361 pKa = 9.02TSWDD365 pKa = 3.5DD366 pKa = 3.56QGFLTIVTTDD376 pKa = 3.62ASWLTAARR384 pKa = 11.84SYY386 pKa = 11.35DD387 pKa = 3.55EE388 pKa = 4.32QGFLITPSAPTIDD401 pKa = 3.4TGLPVPTTGPITTNSEE417 pKa = 3.79PDD419 pKa = 3.5PASTAGAVTAAPTTTFEE436 pKa = 4.1TAPNEE441 pKa = 4.09EE442 pKa = 4.23SQVPSNEE449 pKa = 4.28IITADD454 pKa = 3.45ATTTAIPSADD464 pKa = 3.35GTRR467 pKa = 11.84VAEE470 pKa = 4.35GDD472 pKa = 3.85SDD474 pKa = 4.42VSTADD479 pKa = 3.44STPVTSTTTGSEE491 pKa = 4.36SVPSSSSEE499 pKa = 4.07SSLSTATQTTGTDD512 pKa = 3.51AQTSATPSSDD522 pKa = 2.97SSNSPSATATSASEE536 pKa = 4.17VSATSTSLSSTKK548 pKa = 10.58EE549 pKa = 3.7SITRR553 pKa = 11.84SNSPTSTRR561 pKa = 11.84EE562 pKa = 3.62NTTLATSTEE571 pKa = 4.49SGASTTSASGTPPAPNGVGRR591 pKa = 11.84QGGMQPLGALLLGIVVLVLII611 pKa = 5.04
MM1 pKa = 7.61FSTLSISPSMLSISALTLTSLLVPSAAQTFTEE33 pKa = 4.86CNPLTQGGCPPDD45 pKa = 3.94PALGRR50 pKa = 11.84SVSVDD55 pKa = 3.85FTSGPSSEE63 pKa = 4.15FSAVGSTSYY72 pKa = 10.72TSDD75 pKa = 2.85GAAFTVARR83 pKa = 11.84SGDD86 pKa = 3.84SPSITSEE93 pKa = 3.56WYY95 pKa = 9.88IMFGHH100 pKa = 6.54VDD102 pKa = 4.14FVVKK106 pKa = 10.39AAPGTGIVSAAILQSDD122 pKa = 4.62CLDD125 pKa = 4.28EE126 pKa = 6.5IDD128 pKa = 4.05WEE130 pKa = 4.29WLGGDD135 pKa = 3.56NSQAQSNYY143 pKa = 8.23FGKK146 pKa = 11.16GITTTGYY153 pKa = 10.5NRR155 pKa = 11.84GAFHH159 pKa = 6.95GASNNHH165 pKa = 6.09DD166 pKa = 3.5QFHH169 pKa = 6.7TYY171 pKa = 10.38SIDD174 pKa = 2.94WTAEE178 pKa = 3.89QIVWSIDD185 pKa = 3.21GTTVRR190 pKa = 11.84AMSQAQAEE198 pKa = 4.46PGQYY202 pKa = 9.13PQTPMYY208 pKa = 11.08VKK210 pKa = 9.77MGAWAGGDD218 pKa = 3.55PANAPGTIEE227 pKa = 3.88WAGGYY232 pKa = 8.06TDD234 pKa = 4.61YY235 pKa = 11.44SQGPFTMYY243 pKa = 10.22VKK245 pKa = 10.46SVQVTDD251 pKa = 3.63YY252 pKa = 10.25STGSEE257 pKa = 4.08YY258 pKa = 10.36TYY260 pKa = 10.84SGSSGTWQSIQSNGGSINPEE280 pKa = 3.88GKK282 pKa = 8.6GTPVSPDD289 pKa = 2.93SAPAVTSVVPDD300 pKa = 3.41NGAPLGFGNNANRR313 pKa = 11.84EE314 pKa = 4.33SIAATRR320 pKa = 11.84TGYY323 pKa = 9.74PWTGTATASIATASTHH339 pKa = 4.52QTSDD343 pKa = 3.46SSWPSAPASDD353 pKa = 4.56SPSSPIVYY361 pKa = 9.02TSWDD365 pKa = 3.5DD366 pKa = 3.56QGFLTIVTTDD376 pKa = 3.62ASWLTAARR384 pKa = 11.84SYY386 pKa = 11.35DD387 pKa = 3.55EE388 pKa = 4.32QGFLITPSAPTIDD401 pKa = 3.4TGLPVPTTGPITTNSEE417 pKa = 3.79PDD419 pKa = 3.5PASTAGAVTAAPTTTFEE436 pKa = 4.1TAPNEE441 pKa = 4.09EE442 pKa = 4.23SQVPSNEE449 pKa = 4.28IITADD454 pKa = 3.45ATTTAIPSADD464 pKa = 3.35GTRR467 pKa = 11.84VAEE470 pKa = 4.35GDD472 pKa = 3.85SDD474 pKa = 4.42VSTADD479 pKa = 3.44STPVTSTTTGSEE491 pKa = 4.36SVPSSSSEE499 pKa = 4.07SSLSTATQTTGTDD512 pKa = 3.51AQTSATPSSDD522 pKa = 2.97SSNSPSATATSASEE536 pKa = 4.17VSATSTSLSSTKK548 pKa = 10.58EE549 pKa = 3.7SITRR553 pKa = 11.84SNSPTSTRR561 pKa = 11.84EE562 pKa = 3.62NTTLATSTEE571 pKa = 4.49SGASTTSASGTPPAPNGVGRR591 pKa = 11.84QGGMQPLGALLLGIVVLVLII611 pKa = 5.04
Molecular weight: 62.35 kDa
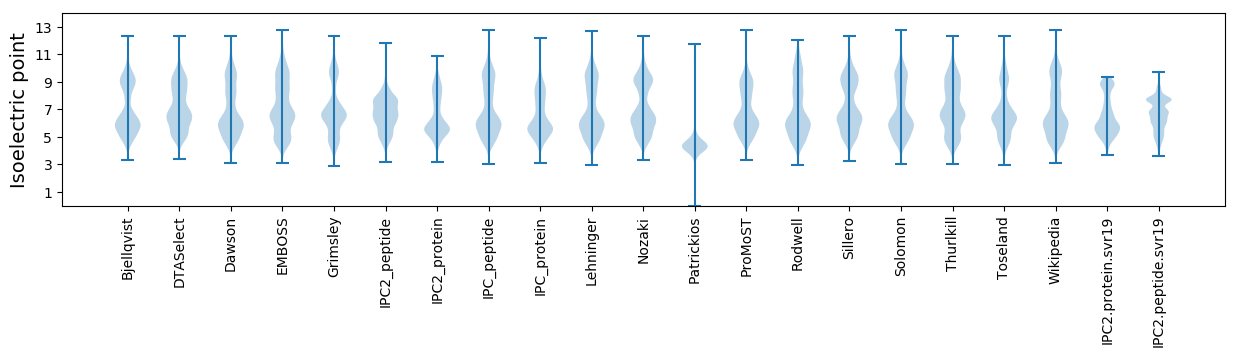
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D1WS74|A0A0D1WS74_EXOME Transcription initiation factor IIE subunit alpha OS=Exophiala mesophila OX=212818 GN=PV10_06482 PE=3 SV=1
MM1 pKa = 6.79QVCDD5 pKa = 3.79RR6 pKa = 11.84VLGSWSNVVQGHH18 pKa = 5.37IRR20 pKa = 11.84LFRR23 pKa = 11.84RR24 pKa = 11.84GMSVMVYY31 pKa = 8.79QVFPSARR38 pKa = 11.84IMLALRR44 pKa = 11.84LAFDD48 pKa = 3.67MLKK51 pKa = 10.72AADD54 pKa = 4.35FPPAVRR60 pKa = 11.84GLSTLLSRR68 pKa = 11.84RR69 pKa = 11.84TAISIAKK76 pKa = 8.1EE77 pKa = 4.11LPHH80 pKa = 6.74SNPFVGSLTALASWWSGSALRR101 pKa = 11.84IFAILTPDD109 pKa = 3.59TLQVEE114 pKa = 4.95ACSMTAGLAPWGIVEE129 pKa = 4.97ALQILPRR136 pKa = 11.84TQASPRR142 pKa = 11.84YY143 pKa = 8.85EE144 pKa = 4.41SNIHH148 pKa = 6.37LNVILKK154 pKa = 10.23
MM1 pKa = 6.79QVCDD5 pKa = 3.79RR6 pKa = 11.84VLGSWSNVVQGHH18 pKa = 5.37IRR20 pKa = 11.84LFRR23 pKa = 11.84RR24 pKa = 11.84GMSVMVYY31 pKa = 8.79QVFPSARR38 pKa = 11.84IMLALRR44 pKa = 11.84LAFDD48 pKa = 3.67MLKK51 pKa = 10.72AADD54 pKa = 4.35FPPAVRR60 pKa = 11.84GLSTLLSRR68 pKa = 11.84RR69 pKa = 11.84TAISIAKK76 pKa = 8.1EE77 pKa = 4.11LPHH80 pKa = 6.74SNPFVGSLTALASWWSGSALRR101 pKa = 11.84IFAILTPDD109 pKa = 3.59TLQVEE114 pKa = 4.95ACSMTAGLAPWGIVEE129 pKa = 4.97ALQILPRR136 pKa = 11.84TQASPRR142 pKa = 11.84YY143 pKa = 8.85EE144 pKa = 4.41SNIHH148 pKa = 6.37LNVILKK154 pKa = 10.23
Molecular weight: 16.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5129384 |
49 |
5074 |
512.9 |
56.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.371 ± 0.017 | 1.111 ± 0.008 |
5.888 ± 0.016 | 5.845 ± 0.02 |
3.659 ± 0.013 | 6.626 ± 0.024 |
2.517 ± 0.012 | 4.954 ± 0.015 |
4.769 ± 0.023 | 8.911 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.097 ± 0.009 | 3.73 ± 0.011 |
6.192 ± 0.024 | 4.307 ± 0.015 |
6.097 ± 0.019 | 8.684 ± 0.028 |
6.191 ± 0.02 | 5.992 ± 0.016 |
1.383 ± 0.008 | 2.675 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
