
Poseidonocella sedimentorum
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Poseidonocella
Average proteome isoelectric point is 6.09
Get precalculated fractions of proteins
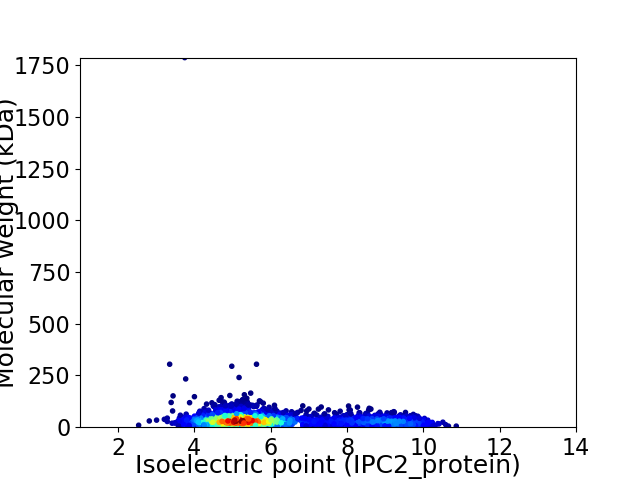
Virtual 2D-PAGE plot for 3242 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I6DGX3|A0A1I6DGX3_9RHOB Peptidyl-prolyl cis-trans isomerase OS=Poseidonocella sedimentorum OX=871652 GN=SAMN04515673_103218 PE=3 SV=1
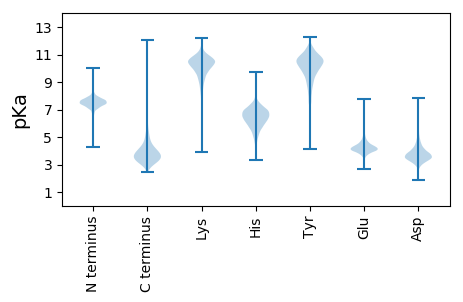
MM1 pKa = 7.16NRR3 pKa = 11.84IFVSGIALAASAGMASAGGIDD24 pKa = 4.7RR25 pKa = 11.84SGQSIGLIFEE35 pKa = 4.59EE36 pKa = 4.72GNYY39 pKa = 10.19AQLSFGSVSPDD50 pKa = 2.95VSGAQVIAVPTLAGVIPAGSEE71 pKa = 3.93SGNMSGSYY79 pKa = 8.17TQFSLGFKK87 pKa = 9.41QAYY90 pKa = 10.15DD91 pKa = 3.57NGLEE95 pKa = 3.99MAFILDD101 pKa = 3.73QPFGADD107 pKa = 3.01VEE109 pKa = 4.58YY110 pKa = 10.35PDD112 pKa = 5.61DD113 pKa = 3.71EE114 pKa = 5.15TYY116 pKa = 10.59FAAGSTAEE124 pKa = 4.36LSSTAITGIVKK135 pKa = 10.39YY136 pKa = 10.29SLPNNVSVFGGLRR149 pKa = 11.84YY150 pKa = 9.7QSLEE154 pKa = 3.71AKK156 pKa = 10.21AYY158 pKa = 9.54IPFVTASAGPTAGVPYY174 pKa = 10.19EE175 pKa = 4.09VDD177 pKa = 3.69GEE179 pKa = 4.27KK180 pKa = 10.64DD181 pKa = 2.74WGLGYY186 pKa = 10.91LLGVAYY192 pKa = 8.29EE193 pKa = 4.42RR194 pKa = 11.84PDD196 pKa = 2.94IALRR200 pKa = 11.84VALTYY205 pKa = 11.05NSAIDD210 pKa = 4.1HH211 pKa = 6.72EE212 pKa = 4.86IDD214 pKa = 3.36TYY216 pKa = 9.88EE217 pKa = 4.17TSALNPGGLDD227 pKa = 3.12TVTDD231 pKa = 3.64VSTPQSLNLEE241 pKa = 4.28FQSGIAADD249 pKa = 3.76TLLFGSVRR257 pKa = 11.84WVDD260 pKa = 2.94WSEE263 pKa = 4.06FDD265 pKa = 5.73ISPQDD270 pKa = 3.43YY271 pKa = 10.09DD272 pKa = 3.65ALLDD276 pKa = 4.07FSRR279 pKa = 11.84GAGDD283 pKa = 3.72YY284 pKa = 11.23SLVSYY289 pKa = 11.32DD290 pKa = 5.71DD291 pKa = 4.03DD292 pKa = 3.91TTSYY296 pKa = 11.62SLGLGRR302 pKa = 11.84RR303 pKa = 11.84INEE306 pKa = 3.84TWSAAVTVGYY316 pKa = 8.24EE317 pKa = 3.75ASAGGFASNLGPTDD331 pKa = 2.96GRR333 pKa = 11.84TSIGLAATYY342 pKa = 10.2TDD344 pKa = 3.94GPMKK348 pKa = 9.81ITGGVSYY355 pKa = 10.97VRR357 pKa = 11.84IGDD360 pKa = 3.96AEE362 pKa = 4.31TTLDD366 pKa = 3.6GTSQATEE373 pKa = 3.88FDD375 pKa = 4.43DD376 pKa = 4.63NSAIGWGLRR385 pKa = 11.84IGYY388 pKa = 9.77AFF390 pKa = 4.33
MM1 pKa = 7.16NRR3 pKa = 11.84IFVSGIALAASAGMASAGGIDD24 pKa = 4.7RR25 pKa = 11.84SGQSIGLIFEE35 pKa = 4.59EE36 pKa = 4.72GNYY39 pKa = 10.19AQLSFGSVSPDD50 pKa = 2.95VSGAQVIAVPTLAGVIPAGSEE71 pKa = 3.93SGNMSGSYY79 pKa = 8.17TQFSLGFKK87 pKa = 9.41QAYY90 pKa = 10.15DD91 pKa = 3.57NGLEE95 pKa = 3.99MAFILDD101 pKa = 3.73QPFGADD107 pKa = 3.01VEE109 pKa = 4.58YY110 pKa = 10.35PDD112 pKa = 5.61DD113 pKa = 3.71EE114 pKa = 5.15TYY116 pKa = 10.59FAAGSTAEE124 pKa = 4.36LSSTAITGIVKK135 pKa = 10.39YY136 pKa = 10.29SLPNNVSVFGGLRR149 pKa = 11.84YY150 pKa = 9.7QSLEE154 pKa = 3.71AKK156 pKa = 10.21AYY158 pKa = 9.54IPFVTASAGPTAGVPYY174 pKa = 10.19EE175 pKa = 4.09VDD177 pKa = 3.69GEE179 pKa = 4.27KK180 pKa = 10.64DD181 pKa = 2.74WGLGYY186 pKa = 10.91LLGVAYY192 pKa = 8.29EE193 pKa = 4.42RR194 pKa = 11.84PDD196 pKa = 2.94IALRR200 pKa = 11.84VALTYY205 pKa = 11.05NSAIDD210 pKa = 4.1HH211 pKa = 6.72EE212 pKa = 4.86IDD214 pKa = 3.36TYY216 pKa = 9.88EE217 pKa = 4.17TSALNPGGLDD227 pKa = 3.12TVTDD231 pKa = 3.64VSTPQSLNLEE241 pKa = 4.28FQSGIAADD249 pKa = 3.76TLLFGSVRR257 pKa = 11.84WVDD260 pKa = 2.94WSEE263 pKa = 4.06FDD265 pKa = 5.73ISPQDD270 pKa = 3.43YY271 pKa = 10.09DD272 pKa = 3.65ALLDD276 pKa = 4.07FSRR279 pKa = 11.84GAGDD283 pKa = 3.72YY284 pKa = 11.23SLVSYY289 pKa = 11.32DD290 pKa = 5.71DD291 pKa = 4.03DD292 pKa = 3.91TTSYY296 pKa = 11.62SLGLGRR302 pKa = 11.84RR303 pKa = 11.84INEE306 pKa = 3.84TWSAAVTVGYY316 pKa = 8.24EE317 pKa = 3.75ASAGGFASNLGPTDD331 pKa = 2.96GRR333 pKa = 11.84TSIGLAATYY342 pKa = 10.2TDD344 pKa = 3.94GPMKK348 pKa = 9.81ITGGVSYY355 pKa = 10.97VRR357 pKa = 11.84IGDD360 pKa = 3.96AEE362 pKa = 4.31TTLDD366 pKa = 3.6GTSQATEE373 pKa = 3.88FDD375 pKa = 4.43DD376 pKa = 4.63NSAIGWGLRR385 pKa = 11.84IGYY388 pKa = 9.77AFF390 pKa = 4.33
Molecular weight: 40.9 kDa
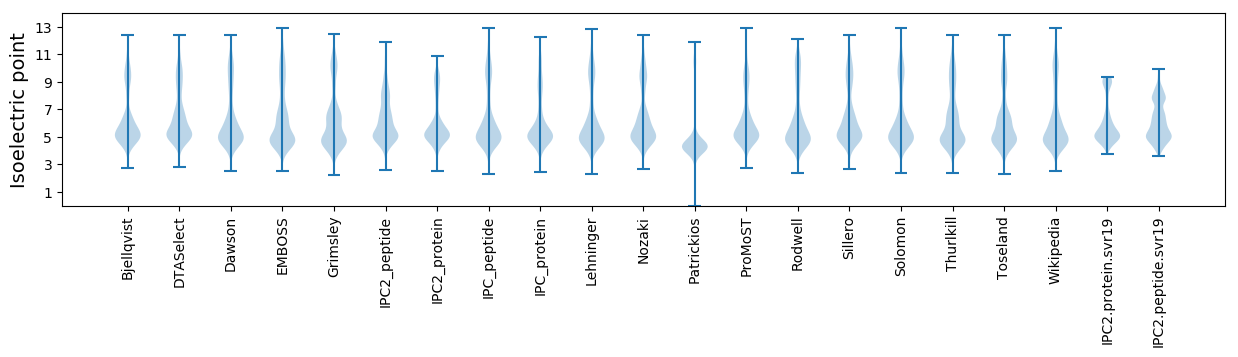
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I6DHP4|A0A1I6DHP4_9RHOB Uncharacterized protein OS=Poseidonocella sedimentorum OX=871652 GN=SAMN04515673_103276 PE=4 SV=1
MM1 pKa = 7.09TRR3 pKa = 11.84YY4 pKa = 9.62PPTAWRR10 pKa = 11.84ALRR13 pKa = 11.84PVKK16 pKa = 9.15TARR19 pKa = 11.84ALAVLAALCLAAPAWAAGAKK39 pKa = 10.0KK40 pKa = 10.05PEE42 pKa = 4.38APAGPVNSTPACQVMDD58 pKa = 5.3KK59 pKa = 10.68ILALDD64 pKa = 4.01PLSHH68 pKa = 6.42KK69 pKa = 10.41VAVGIDD75 pKa = 3.47HH76 pKa = 6.85FQLIDD81 pKa = 3.29QYY83 pKa = 9.84RR84 pKa = 11.84TKK86 pKa = 10.52IEE88 pKa = 3.75IARR91 pKa = 11.84QSLRR95 pKa = 11.84ALPASVSVPDD105 pKa = 3.66RR106 pKa = 11.84EE107 pKa = 3.97RR108 pKa = 11.84LQDD111 pKa = 3.62HH112 pKa = 5.46VTAHH116 pKa = 6.97ADD118 pKa = 3.32VAEE121 pKa = 4.37TVAINGSSPAAALITSEE138 pKa = 4.08PFLRR142 pKa = 11.84RR143 pKa = 11.84ALQVEE148 pKa = 4.43ADD150 pKa = 4.06HH151 pKa = 6.99KK152 pKa = 11.46ALGCLSPSWRR162 pKa = 11.84DD163 pKa = 3.1RR164 pKa = 11.84LSFLTDD170 pKa = 3.54LFKK173 pKa = 10.58PVQRR177 pKa = 11.84IWAHH181 pKa = 4.32EE182 pKa = 3.92RR183 pKa = 11.84VRR185 pKa = 11.84PFILWGICFSLVVWARR201 pKa = 11.84RR202 pKa = 11.84RR203 pKa = 11.84QKK205 pKa = 10.5RR206 pKa = 11.84YY207 pKa = 8.76QALNALPDD215 pKa = 3.56YY216 pKa = 10.3DD217 pKa = 4.24ARR219 pKa = 11.84EE220 pKa = 4.18APRR223 pKa = 11.84VNCSLPAQVHH233 pKa = 5.84IEE235 pKa = 3.81ADD237 pKa = 4.41GIPIRR242 pKa = 11.84ILDD245 pKa = 3.78ISRR248 pKa = 11.84KK249 pKa = 7.11GARR252 pKa = 11.84FALSRR257 pKa = 11.84PVEE260 pKa = 4.43LPDD263 pKa = 4.73EE264 pKa = 4.11IHH266 pKa = 6.47IALPAEE272 pKa = 4.17IRR274 pKa = 11.84RR275 pKa = 11.84TALLRR280 pKa = 11.84RR281 pKa = 11.84MRR283 pKa = 11.84ARR285 pKa = 11.84HH286 pKa = 5.24GAVMFLPRR294 pKa = 11.84LTDD297 pKa = 3.3VEE299 pKa = 4.22FAKK302 pKa = 10.68VLRR305 pKa = 11.84HH306 pKa = 6.01SDD308 pKa = 3.54LPVASRR314 pKa = 11.84ALPPAKK320 pKa = 9.69PPRR323 pKa = 11.84GAALAGLLGWLPQGRR338 pKa = 11.84RR339 pKa = 11.84SKK341 pKa = 11.22AA342 pKa = 3.05
MM1 pKa = 7.09TRR3 pKa = 11.84YY4 pKa = 9.62PPTAWRR10 pKa = 11.84ALRR13 pKa = 11.84PVKK16 pKa = 9.15TARR19 pKa = 11.84ALAVLAALCLAAPAWAAGAKK39 pKa = 10.0KK40 pKa = 10.05PEE42 pKa = 4.38APAGPVNSTPACQVMDD58 pKa = 5.3KK59 pKa = 10.68ILALDD64 pKa = 4.01PLSHH68 pKa = 6.42KK69 pKa = 10.41VAVGIDD75 pKa = 3.47HH76 pKa = 6.85FQLIDD81 pKa = 3.29QYY83 pKa = 9.84RR84 pKa = 11.84TKK86 pKa = 10.52IEE88 pKa = 3.75IARR91 pKa = 11.84QSLRR95 pKa = 11.84ALPASVSVPDD105 pKa = 3.66RR106 pKa = 11.84EE107 pKa = 3.97RR108 pKa = 11.84LQDD111 pKa = 3.62HH112 pKa = 5.46VTAHH116 pKa = 6.97ADD118 pKa = 3.32VAEE121 pKa = 4.37TVAINGSSPAAALITSEE138 pKa = 4.08PFLRR142 pKa = 11.84RR143 pKa = 11.84ALQVEE148 pKa = 4.43ADD150 pKa = 4.06HH151 pKa = 6.99KK152 pKa = 11.46ALGCLSPSWRR162 pKa = 11.84DD163 pKa = 3.1RR164 pKa = 11.84LSFLTDD170 pKa = 3.54LFKK173 pKa = 10.58PVQRR177 pKa = 11.84IWAHH181 pKa = 4.32EE182 pKa = 3.92RR183 pKa = 11.84VRR185 pKa = 11.84PFILWGICFSLVVWARR201 pKa = 11.84RR202 pKa = 11.84RR203 pKa = 11.84QKK205 pKa = 10.5RR206 pKa = 11.84YY207 pKa = 8.76QALNALPDD215 pKa = 3.56YY216 pKa = 10.3DD217 pKa = 4.24ARR219 pKa = 11.84EE220 pKa = 4.18APRR223 pKa = 11.84VNCSLPAQVHH233 pKa = 5.84IEE235 pKa = 3.81ADD237 pKa = 4.41GIPIRR242 pKa = 11.84ILDD245 pKa = 3.78ISRR248 pKa = 11.84KK249 pKa = 7.11GARR252 pKa = 11.84FALSRR257 pKa = 11.84PVEE260 pKa = 4.43LPDD263 pKa = 4.73EE264 pKa = 4.11IHH266 pKa = 6.47IALPAEE272 pKa = 4.17IRR274 pKa = 11.84RR275 pKa = 11.84TALLRR280 pKa = 11.84RR281 pKa = 11.84MRR283 pKa = 11.84ARR285 pKa = 11.84HH286 pKa = 5.24GAVMFLPRR294 pKa = 11.84LTDD297 pKa = 3.3VEE299 pKa = 4.22FAKK302 pKa = 10.68VLRR305 pKa = 11.84HH306 pKa = 6.01SDD308 pKa = 3.54LPVASRR314 pKa = 11.84ALPPAKK320 pKa = 9.69PPRR323 pKa = 11.84GAALAGLLGWLPQGRR338 pKa = 11.84RR339 pKa = 11.84SKK341 pKa = 11.22AA342 pKa = 3.05
Molecular weight: 37.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1066642 |
30 |
17790 |
329.0 |
35.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.109 ± 0.071 | 0.84 ± 0.02 |
6.017 ± 0.066 | 6.197 ± 0.042 |
3.671 ± 0.033 | 9.017 ± 0.083 |
1.949 ± 0.029 | 5.087 ± 0.03 |
2.686 ± 0.05 | 10.331 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.545 ± 0.037 | 2.285 ± 0.023 |
5.228 ± 0.059 | 2.908 ± 0.023 |
7.073 ± 0.072 | 5.199 ± 0.056 |
5.425 ± 0.087 | 6.989 ± 0.034 |
1.324 ± 0.019 | 2.12 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
