
Costertonia aggregata
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Costertonia
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
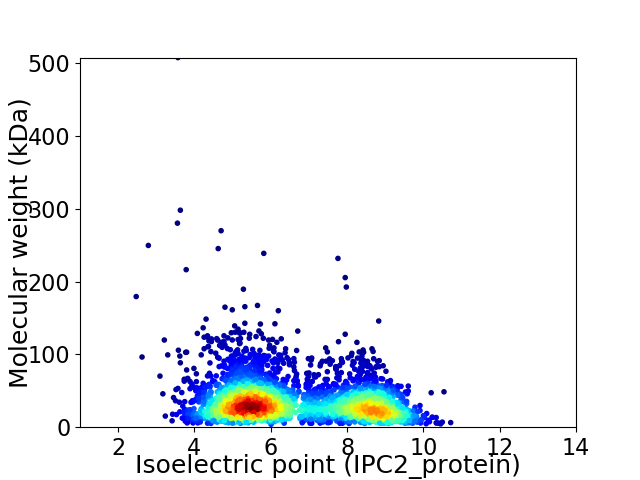
Virtual 2D-PAGE plot for 3525 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A7H9ALA8|A0A7H9ALA8_9FLAO Preprotein translocase subunit SecE OS=Costertonia aggregata OX=343403 GN=secE PE=4 SV=1
MM1 pKa = 7.56TIISKK6 pKa = 10.7NKK8 pKa = 9.18TILLFLFILSSIGYY22 pKa = 8.24GQTTIWEE29 pKa = 4.28EE30 pKa = 3.97DD31 pKa = 3.48FEE33 pKa = 5.6SYY35 pKa = 11.76GNGTSTGAGSGASTATWSTNDD56 pKa = 2.65GDD58 pKa = 3.74IDD60 pKa = 3.67VRR62 pKa = 11.84TISGNKK68 pKa = 7.69VLRR71 pKa = 11.84GSDD74 pKa = 3.29TRR76 pKa = 11.84NTSARR81 pKa = 11.84WTTNPIDD88 pKa = 3.2ISGYY92 pKa = 7.4TNVSFSLDD100 pKa = 3.45ADD102 pKa = 3.76SGGGLDD108 pKa = 3.64SGVDD112 pKa = 3.31VFRR115 pKa = 11.84IQYY118 pKa = 10.05RR119 pKa = 11.84IDD121 pKa = 3.41GGSYY125 pKa = 9.93IEE127 pKa = 4.76IEE129 pKa = 4.28NTSGDD134 pKa = 3.83TSPSEE139 pKa = 4.58PIQPSYY145 pKa = 10.95GVSGLSGSTLEE156 pKa = 5.44FRR158 pKa = 11.84ITMYY162 pKa = 9.59NTTGAEE168 pKa = 4.5FYY170 pKa = 10.69DD171 pKa = 3.46IDD173 pKa = 4.83NILVQGTLPTPDD185 pKa = 4.43CSTISSFPGSFIASNTTLNSNLSGFSSGIITDD217 pKa = 3.62VNVSLNITHH226 pKa = 6.92TWNADD231 pKa = 3.1LDD233 pKa = 3.77ISLISPSGTTVVLSSDD249 pKa = 3.43NGGSGDD255 pKa = 3.63NYY257 pKa = 10.67VNTFFDD263 pKa = 4.22DD264 pKa = 3.61SGSINIISGSSPFTGTFRR282 pKa = 11.84PEE284 pKa = 3.78GNLSDD289 pKa = 4.89FNGEE293 pKa = 4.08NANGTWVLRR302 pKa = 11.84VFDD305 pKa = 4.41DD306 pKa = 3.59TGGDD310 pKa = 3.28TGTLDD315 pKa = 3.44GFSIEE320 pKa = 3.9ICTNSSTDD328 pKa = 3.59LAVSKK333 pKa = 9.54TVDD336 pKa = 3.74DD337 pKa = 4.06NTPSEE342 pKa = 4.22GSNIVYY348 pKa = 8.79TLSVEE353 pKa = 4.33NNGPLDD359 pKa = 3.78ATNVTLTDD367 pKa = 3.52VLPAGVSFVSDD378 pKa = 3.33DD379 pKa = 3.38GAYY382 pKa = 10.49NSSTGVWSVGSLNNGATAILNITASVDD409 pKa = 3.09GSTSGSTITNTITSISADD427 pKa = 3.43QTDD430 pKa = 4.15SNLTADD436 pKa = 4.94DD437 pKa = 4.24LSEE440 pKa = 4.91SIVPSVDD447 pKa = 3.11QPPVVTVTGNQIYY460 pKa = 10.39CPLTSLSIAEE470 pKa = 4.77TISISDD476 pKa = 4.21PDD478 pKa = 4.23DD479 pKa = 3.52TTTDD483 pKa = 2.9AVYY486 pKa = 10.12IQISSGYY493 pKa = 10.05VNGEE497 pKa = 4.16DD498 pKa = 5.16LLTLTGTHH506 pKa = 6.76PNITATWDD514 pKa = 4.03AIEE517 pKa = 5.68GEE519 pKa = 4.47LTLQGPTTYY528 pKa = 10.79SEE530 pKa = 4.57FEE532 pKa = 4.15TAVLAVEE539 pKa = 4.46YY540 pKa = 10.05SSSASAPTGIKK551 pKa = 9.89QFSITPGSANFLPPTGHH568 pKa = 6.19YY569 pKa = 10.8YY570 pKa = 10.79EE571 pKa = 5.65FISDD575 pKa = 5.56LGITWTDD582 pKa = 3.07ARR584 pKa = 11.84DD585 pKa = 3.28AAALRR590 pKa = 11.84TYY592 pKa = 10.69FGLQGYY598 pKa = 8.27LATLTTQAEE607 pKa = 4.27ADD609 pKa = 3.88FSGTQAVGVGWIGASDD625 pKa = 3.68AANEE629 pKa = 4.22GDD631 pKa = 3.73WRR633 pKa = 11.84WVTGPEE639 pKa = 3.71AGTPFWSGTSTGTTVAPTNFAYY661 pKa = 9.81WNGGEE666 pKa = 4.26PNQSGNEE673 pKa = 4.05DD674 pKa = 3.65YY675 pKa = 11.83AHH677 pKa = 6.34ITDD680 pKa = 4.1PSVVRR685 pKa = 11.84GSGGLGSWNDD695 pKa = 3.57LSNTGASSGPYY706 pKa = 8.87QPQGYY711 pKa = 8.11VVEE714 pKa = 4.51YY715 pKa = 9.83GGSPGDD721 pKa = 3.4PVLNITGVTTITVDD735 pKa = 3.75NINPTASNPSPVTVFCPSDD754 pKa = 3.59VPTFDD759 pKa = 3.06ISVVTDD765 pKa = 3.34EE766 pKa = 5.81ADD768 pKa = 3.34NCTTNPLVTFVSDD781 pKa = 3.39VSNGGSNPEE790 pKa = 4.3IITRR794 pKa = 11.84TYY796 pKa = 10.41RR797 pKa = 11.84ISDD800 pKa = 3.51DD801 pKa = 3.37MGNFIDD807 pKa = 3.91VAQTITVDD815 pKa = 4.41PINIDD820 pKa = 3.79TQPLDD825 pKa = 2.91QTTFVNNNASFSVSINNVDD844 pKa = 3.53TYY846 pKa = 10.59QWQVSIDD853 pKa = 3.39GGTNFIDD860 pKa = 3.38ISDD863 pKa = 3.76GTEE866 pKa = 3.72YY867 pKa = 10.54TGTSTSTLTVIAPRR881 pKa = 11.84VDD883 pKa = 3.12KK884 pKa = 11.27NNYY887 pKa = 7.81RR888 pKa = 11.84YY889 pKa = 9.55RR890 pKa = 11.84VAASNSGSGCSMVISNNALLSVRR913 pKa = 11.84VSSVITNRR921 pKa = 11.84RR922 pKa = 11.84ITYY925 pKa = 9.44RR926 pKa = 11.84VKK928 pKa = 11.07NNN930 pKa = 3.07
MM1 pKa = 7.56TIISKK6 pKa = 10.7NKK8 pKa = 9.18TILLFLFILSSIGYY22 pKa = 8.24GQTTIWEE29 pKa = 4.28EE30 pKa = 3.97DD31 pKa = 3.48FEE33 pKa = 5.6SYY35 pKa = 11.76GNGTSTGAGSGASTATWSTNDD56 pKa = 2.65GDD58 pKa = 3.74IDD60 pKa = 3.67VRR62 pKa = 11.84TISGNKK68 pKa = 7.69VLRR71 pKa = 11.84GSDD74 pKa = 3.29TRR76 pKa = 11.84NTSARR81 pKa = 11.84WTTNPIDD88 pKa = 3.2ISGYY92 pKa = 7.4TNVSFSLDD100 pKa = 3.45ADD102 pKa = 3.76SGGGLDD108 pKa = 3.64SGVDD112 pKa = 3.31VFRR115 pKa = 11.84IQYY118 pKa = 10.05RR119 pKa = 11.84IDD121 pKa = 3.41GGSYY125 pKa = 9.93IEE127 pKa = 4.76IEE129 pKa = 4.28NTSGDD134 pKa = 3.83TSPSEE139 pKa = 4.58PIQPSYY145 pKa = 10.95GVSGLSGSTLEE156 pKa = 5.44FRR158 pKa = 11.84ITMYY162 pKa = 9.59NTTGAEE168 pKa = 4.5FYY170 pKa = 10.69DD171 pKa = 3.46IDD173 pKa = 4.83NILVQGTLPTPDD185 pKa = 4.43CSTISSFPGSFIASNTTLNSNLSGFSSGIITDD217 pKa = 3.62VNVSLNITHH226 pKa = 6.92TWNADD231 pKa = 3.1LDD233 pKa = 3.77ISLISPSGTTVVLSSDD249 pKa = 3.43NGGSGDD255 pKa = 3.63NYY257 pKa = 10.67VNTFFDD263 pKa = 4.22DD264 pKa = 3.61SGSINIISGSSPFTGTFRR282 pKa = 11.84PEE284 pKa = 3.78GNLSDD289 pKa = 4.89FNGEE293 pKa = 4.08NANGTWVLRR302 pKa = 11.84VFDD305 pKa = 4.41DD306 pKa = 3.59TGGDD310 pKa = 3.28TGTLDD315 pKa = 3.44GFSIEE320 pKa = 3.9ICTNSSTDD328 pKa = 3.59LAVSKK333 pKa = 9.54TVDD336 pKa = 3.74DD337 pKa = 4.06NTPSEE342 pKa = 4.22GSNIVYY348 pKa = 8.79TLSVEE353 pKa = 4.33NNGPLDD359 pKa = 3.78ATNVTLTDD367 pKa = 3.52VLPAGVSFVSDD378 pKa = 3.33DD379 pKa = 3.38GAYY382 pKa = 10.49NSSTGVWSVGSLNNGATAILNITASVDD409 pKa = 3.09GSTSGSTITNTITSISADD427 pKa = 3.43QTDD430 pKa = 4.15SNLTADD436 pKa = 4.94DD437 pKa = 4.24LSEE440 pKa = 4.91SIVPSVDD447 pKa = 3.11QPPVVTVTGNQIYY460 pKa = 10.39CPLTSLSIAEE470 pKa = 4.77TISISDD476 pKa = 4.21PDD478 pKa = 4.23DD479 pKa = 3.52TTTDD483 pKa = 2.9AVYY486 pKa = 10.12IQISSGYY493 pKa = 10.05VNGEE497 pKa = 4.16DD498 pKa = 5.16LLTLTGTHH506 pKa = 6.76PNITATWDD514 pKa = 4.03AIEE517 pKa = 5.68GEE519 pKa = 4.47LTLQGPTTYY528 pKa = 10.79SEE530 pKa = 4.57FEE532 pKa = 4.15TAVLAVEE539 pKa = 4.46YY540 pKa = 10.05SSSASAPTGIKK551 pKa = 9.89QFSITPGSANFLPPTGHH568 pKa = 6.19YY569 pKa = 10.8YY570 pKa = 10.79EE571 pKa = 5.65FISDD575 pKa = 5.56LGITWTDD582 pKa = 3.07ARR584 pKa = 11.84DD585 pKa = 3.28AAALRR590 pKa = 11.84TYY592 pKa = 10.69FGLQGYY598 pKa = 8.27LATLTTQAEE607 pKa = 4.27ADD609 pKa = 3.88FSGTQAVGVGWIGASDD625 pKa = 3.68AANEE629 pKa = 4.22GDD631 pKa = 3.73WRR633 pKa = 11.84WVTGPEE639 pKa = 3.71AGTPFWSGTSTGTTVAPTNFAYY661 pKa = 9.81WNGGEE666 pKa = 4.26PNQSGNEE673 pKa = 4.05DD674 pKa = 3.65YY675 pKa = 11.83AHH677 pKa = 6.34ITDD680 pKa = 4.1PSVVRR685 pKa = 11.84GSGGLGSWNDD695 pKa = 3.57LSNTGASSGPYY706 pKa = 8.87QPQGYY711 pKa = 8.11VVEE714 pKa = 4.51YY715 pKa = 9.83GGSPGDD721 pKa = 3.4PVLNITGVTTITVDD735 pKa = 3.75NINPTASNPSPVTVFCPSDD754 pKa = 3.59VPTFDD759 pKa = 3.06ISVVTDD765 pKa = 3.34EE766 pKa = 5.81ADD768 pKa = 3.34NCTTNPLVTFVSDD781 pKa = 3.39VSNGGSNPEE790 pKa = 4.3IITRR794 pKa = 11.84TYY796 pKa = 10.41RR797 pKa = 11.84ISDD800 pKa = 3.51DD801 pKa = 3.37MGNFIDD807 pKa = 3.91VAQTITVDD815 pKa = 4.41PINIDD820 pKa = 3.79TQPLDD825 pKa = 2.91QTTFVNNNASFSVSINNVDD844 pKa = 3.53TYY846 pKa = 10.59QWQVSIDD853 pKa = 3.39GGTNFIDD860 pKa = 3.38ISDD863 pKa = 3.76GTEE866 pKa = 3.72YY867 pKa = 10.54TGTSTSTLTVIAPRR881 pKa = 11.84VDD883 pKa = 3.12KK884 pKa = 11.27NNYY887 pKa = 7.81RR888 pKa = 11.84YY889 pKa = 9.55RR890 pKa = 11.84VAASNSGSGCSMVISNNALLSVRR913 pKa = 11.84VSSVITNRR921 pKa = 11.84RR922 pKa = 11.84ITYY925 pKa = 9.44RR926 pKa = 11.84VKK928 pKa = 11.07NNN930 pKa = 3.07
Molecular weight: 97.7 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A7H9AKM5|A0A7H9AKM5_9FLAO VOC family protein OS=Costertonia aggregata OX=343403 GN=HYG79_01080 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 10.15RR11 pKa = 11.84KK12 pKa = 9.6RR13 pKa = 11.84KK14 pKa = 8.29NKK16 pKa = 9.34HH17 pKa = 4.03GFRR20 pKa = 11.84EE21 pKa = 4.05RR22 pKa = 11.84MASVNGRR29 pKa = 11.84KK30 pKa = 9.21VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.08GRR40 pKa = 11.84KK41 pKa = 8.3KK42 pKa = 9.83LTVSSEE48 pKa = 3.9PRR50 pKa = 11.84HH51 pKa = 5.77KK52 pKa = 10.61KK53 pKa = 9.84
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 10.15RR11 pKa = 11.84KK12 pKa = 9.6RR13 pKa = 11.84KK14 pKa = 8.29NKK16 pKa = 9.34HH17 pKa = 4.03GFRR20 pKa = 11.84EE21 pKa = 4.05RR22 pKa = 11.84MASVNGRR29 pKa = 11.84KK30 pKa = 9.21VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.08GRR40 pKa = 11.84KK41 pKa = 8.3KK42 pKa = 9.83LTVSSEE48 pKa = 3.9PRR50 pKa = 11.84HH51 pKa = 5.77KK52 pKa = 10.61KK53 pKa = 9.84
Molecular weight: 6.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1168523 |
34 |
4800 |
331.5 |
37.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.411 ± 0.041 | 0.738 ± 0.014 |
5.86 ± 0.045 | 6.575 ± 0.038 |
5.237 ± 0.034 | 6.685 ± 0.055 |
1.783 ± 0.023 | 7.62 ± 0.041 |
7.757 ± 0.068 | 9.261 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.284 ± 0.024 | 5.88 ± 0.039 |
3.474 ± 0.027 | 3.296 ± 0.022 |
3.66 ± 0.028 | 6.299 ± 0.029 |
5.824 ± 0.06 | 6.287 ± 0.032 |
1.091 ± 0.017 | 3.978 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
