
Prauserella rugosa
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Pseudonocardiales; Pseudonocardiaceae; Prauserella
Average proteome isoelectric point is 5.91
Get precalculated fractions of proteins
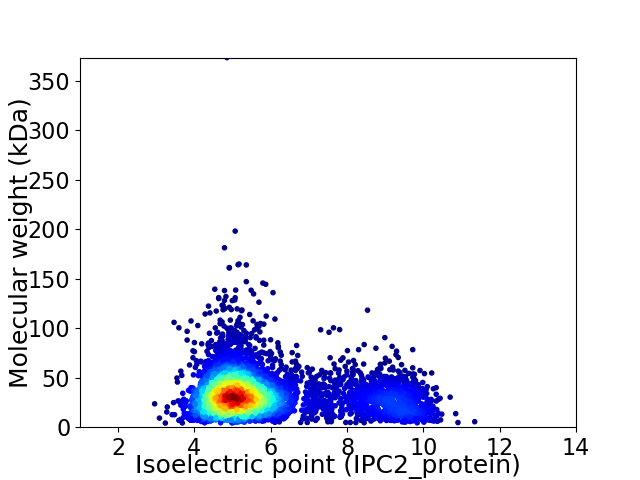
Virtual 2D-PAGE plot for 4886 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
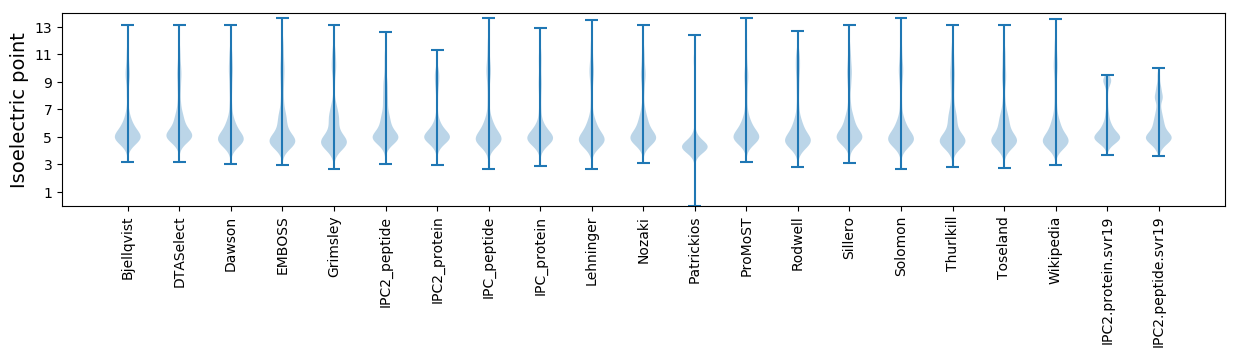
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A660CIE3|A0A660CIE3_9PSEU Uncharacterized protein OS=Prauserella rugosa OX=43354 GN=JD82_03473 PE=4 SV=1
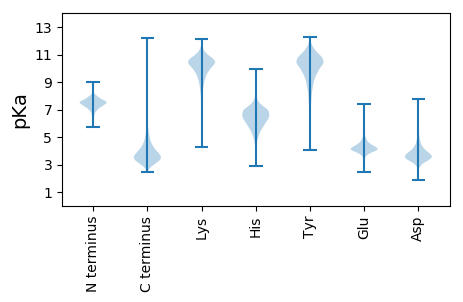
MM1 pKa = 7.75RR2 pKa = 11.84SDD4 pKa = 3.02KK5 pKa = 10.89RR6 pKa = 11.84RR7 pKa = 11.84IGVLAGAGTAAVAVTLSMAGTANAVPQEE35 pKa = 4.11GTVLGAGSATAVEE48 pKa = 4.38GKK50 pKa = 10.65YY51 pKa = 10.23IVVLKK56 pKa = 10.97DD57 pKa = 3.51GVQTTAADD65 pKa = 3.44LGGEE69 pKa = 4.3VNHH72 pKa = 6.6TFSTVLNGYY81 pKa = 8.7SADD84 pKa = 3.52MSEE87 pKa = 4.22ALAEE91 pKa = 3.89RR92 pKa = 11.84VAANPAVDD100 pKa = 3.72YY101 pKa = 11.11VEE103 pKa = 4.59QVQTVHH109 pKa = 6.0TMADD113 pKa = 3.36QTDD116 pKa = 3.97PPSWGLDD123 pKa = 3.18RR124 pKa = 11.84IDD126 pKa = 4.0QAEE129 pKa = 4.32LPLDD133 pKa = 3.61GTYY136 pKa = 9.76TYY138 pKa = 10.17PNEE141 pKa = 4.55GEE143 pKa = 4.47GVTAYY148 pKa = 9.87IIDD151 pKa = 3.76TGIMTSHH158 pKa = 6.44QDD160 pKa = 3.08FGGRR164 pKa = 11.84ATAGFDD170 pKa = 3.57AVDD173 pKa = 3.57SGGDD177 pKa = 3.41AEE179 pKa = 4.85DD180 pKa = 4.54CNGHH184 pKa = 5.09GTHH187 pKa = 6.37VAGTVGGSEE196 pKa = 4.11YY197 pKa = 11.07GVAKK201 pKa = 10.5SADD204 pKa = 3.53LVAVRR209 pKa = 11.84VLNCQGSGTNEE220 pKa = 3.58QVIAGIDD227 pKa = 3.39WVTEE231 pKa = 4.04NASGPSVANMSLGGSASAAIDD252 pKa = 3.58DD253 pKa = 4.28AVQNSIASGVTYY265 pKa = 10.84GLAAGNDD272 pKa = 3.88SGADD276 pKa = 3.39ACNSSPARR284 pKa = 11.84VPEE287 pKa = 4.12GLTVGSTTEE296 pKa = 3.46TDD298 pKa = 3.2ARR300 pKa = 11.84SSFSNIGSCVDD311 pKa = 3.37IFAPGSDD318 pKa = 3.5ITSAWIDD325 pKa = 3.28SDD327 pKa = 3.54TSEE330 pKa = 4.08NTISGTSMATPHH342 pKa = 5.72VVGAAALYY350 pKa = 10.02LAANPDD356 pKa = 3.45ATPAQVGDD364 pKa = 3.8ALVAGGSEE372 pKa = 4.06GVVGNPGSGSPNVLLNVVTDD392 pKa = 4.86GSAPDD397 pKa = 4.65PDD399 pKa = 5.09PEE401 pKa = 4.6PEE403 pKa = 4.78PDD405 pKa = 3.89PGTCEE410 pKa = 4.16AVTNDD415 pKa = 4.09EE416 pKa = 4.04VTAIPDD422 pKa = 3.67AGSVDD427 pKa = 3.64STVTVSGCDD436 pKa = 3.18AASAEE441 pKa = 4.26ATVEE445 pKa = 3.62VDD447 pKa = 2.5ITHH450 pKa = 7.53TYY452 pKa = 10.14RR453 pKa = 11.84GDD455 pKa = 3.28IALDD459 pKa = 4.35LIAPGGAEE467 pKa = 4.0FSLKK471 pKa = 9.82QASFDD476 pKa = 3.57SGTDD480 pKa = 2.79ITEE483 pKa = 4.25TYY485 pKa = 9.78TVDD488 pKa = 3.21LSGVTEE494 pKa = 4.63VDD496 pKa = 3.77GEE498 pKa = 4.34WTLKK502 pKa = 9.15VTDD505 pKa = 3.84SFVYY509 pKa = 9.08DD510 pKa = 3.54TGTLNSWTLSLL521 pKa = 4.75
MM1 pKa = 7.75RR2 pKa = 11.84SDD4 pKa = 3.02KK5 pKa = 10.89RR6 pKa = 11.84RR7 pKa = 11.84IGVLAGAGTAAVAVTLSMAGTANAVPQEE35 pKa = 4.11GTVLGAGSATAVEE48 pKa = 4.38GKK50 pKa = 10.65YY51 pKa = 10.23IVVLKK56 pKa = 10.97DD57 pKa = 3.51GVQTTAADD65 pKa = 3.44LGGEE69 pKa = 4.3VNHH72 pKa = 6.6TFSTVLNGYY81 pKa = 8.7SADD84 pKa = 3.52MSEE87 pKa = 4.22ALAEE91 pKa = 3.89RR92 pKa = 11.84VAANPAVDD100 pKa = 3.72YY101 pKa = 11.11VEE103 pKa = 4.59QVQTVHH109 pKa = 6.0TMADD113 pKa = 3.36QTDD116 pKa = 3.97PPSWGLDD123 pKa = 3.18RR124 pKa = 11.84IDD126 pKa = 4.0QAEE129 pKa = 4.32LPLDD133 pKa = 3.61GTYY136 pKa = 9.76TYY138 pKa = 10.17PNEE141 pKa = 4.55GEE143 pKa = 4.47GVTAYY148 pKa = 9.87IIDD151 pKa = 3.76TGIMTSHH158 pKa = 6.44QDD160 pKa = 3.08FGGRR164 pKa = 11.84ATAGFDD170 pKa = 3.57AVDD173 pKa = 3.57SGGDD177 pKa = 3.41AEE179 pKa = 4.85DD180 pKa = 4.54CNGHH184 pKa = 5.09GTHH187 pKa = 6.37VAGTVGGSEE196 pKa = 4.11YY197 pKa = 11.07GVAKK201 pKa = 10.5SADD204 pKa = 3.53LVAVRR209 pKa = 11.84VLNCQGSGTNEE220 pKa = 3.58QVIAGIDD227 pKa = 3.39WVTEE231 pKa = 4.04NASGPSVANMSLGGSASAAIDD252 pKa = 3.58DD253 pKa = 4.28AVQNSIASGVTYY265 pKa = 10.84GLAAGNDD272 pKa = 3.88SGADD276 pKa = 3.39ACNSSPARR284 pKa = 11.84VPEE287 pKa = 4.12GLTVGSTTEE296 pKa = 3.46TDD298 pKa = 3.2ARR300 pKa = 11.84SSFSNIGSCVDD311 pKa = 3.37IFAPGSDD318 pKa = 3.5ITSAWIDD325 pKa = 3.28SDD327 pKa = 3.54TSEE330 pKa = 4.08NTISGTSMATPHH342 pKa = 5.72VVGAAALYY350 pKa = 10.02LAANPDD356 pKa = 3.45ATPAQVGDD364 pKa = 3.8ALVAGGSEE372 pKa = 4.06GVVGNPGSGSPNVLLNVVTDD392 pKa = 4.86GSAPDD397 pKa = 4.65PDD399 pKa = 5.09PEE401 pKa = 4.6PEE403 pKa = 4.78PDD405 pKa = 3.89PGTCEE410 pKa = 4.16AVTNDD415 pKa = 4.09EE416 pKa = 4.04VTAIPDD422 pKa = 3.67AGSVDD427 pKa = 3.64STVTVSGCDD436 pKa = 3.18AASAEE441 pKa = 4.26ATVEE445 pKa = 3.62VDD447 pKa = 2.5ITHH450 pKa = 7.53TYY452 pKa = 10.14RR453 pKa = 11.84GDD455 pKa = 3.28IALDD459 pKa = 4.35LIAPGGAEE467 pKa = 4.0FSLKK471 pKa = 9.82QASFDD476 pKa = 3.57SGTDD480 pKa = 2.79ITEE483 pKa = 4.25TYY485 pKa = 9.78TVDD488 pKa = 3.21LSGVTEE494 pKa = 4.63VDD496 pKa = 3.77GEE498 pKa = 4.34WTLKK502 pKa = 9.15VTDD505 pKa = 3.84SFVYY509 pKa = 9.08DD510 pKa = 3.54TGTLNSWTLSLL521 pKa = 4.75
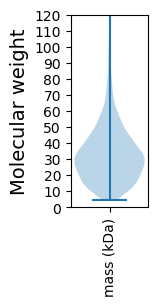
Molecular weight: 52.35 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A660CIX2|A0A660CIX2_9PSEU Glycerol-3-phosphate dehydrogenase OS=Prauserella rugosa OX=43354 GN=JD82_02776 PE=3 SV=1
MM1 pKa = 7.53SKK3 pKa = 10.53GKK5 pKa = 8.66RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84ARR17 pKa = 11.84THH19 pKa = 5.69GFRR22 pKa = 11.84LRR24 pKa = 11.84MRR26 pKa = 11.84TRR28 pKa = 11.84AGRR31 pKa = 11.84AIVAARR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 9.62GRR42 pKa = 11.84QQLSAA47 pKa = 4.05
MM1 pKa = 7.53SKK3 pKa = 10.53GKK5 pKa = 8.66RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84ARR17 pKa = 11.84THH19 pKa = 5.69GFRR22 pKa = 11.84LRR24 pKa = 11.84MRR26 pKa = 11.84TRR28 pKa = 11.84AGRR31 pKa = 11.84AIVAARR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 9.62GRR42 pKa = 11.84QQLSAA47 pKa = 4.05
Molecular weight: 5.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1566181 |
39 |
3507 |
320.5 |
34.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.199 ± 0.051 | 0.728 ± 0.008 |
6.549 ± 0.03 | 6.042 ± 0.03 |
2.721 ± 0.022 | 9.593 ± 0.036 |
2.235 ± 0.016 | 3.367 ± 0.024 |
1.978 ± 0.027 | 9.794 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.818 ± 0.013 | 1.839 ± 0.015 |
5.799 ± 0.03 | 2.759 ± 0.021 |
7.809 ± 0.037 | 5.242 ± 0.023 |
6.12 ± 0.028 | 9.026 ± 0.035 |
1.414 ± 0.017 | 1.968 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
