
Capybara microvirus Cap1_SP_84
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
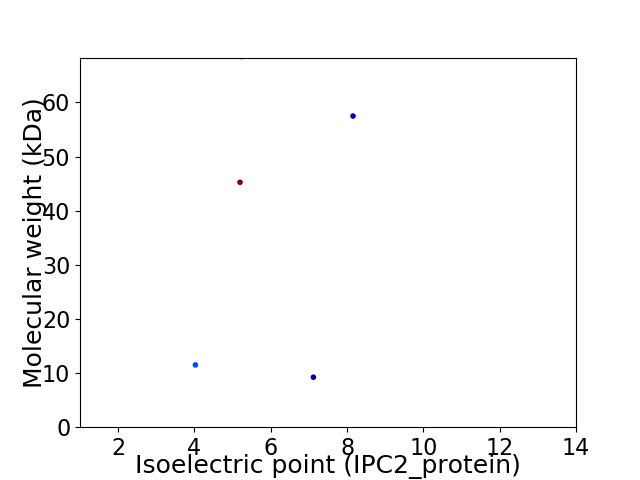
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
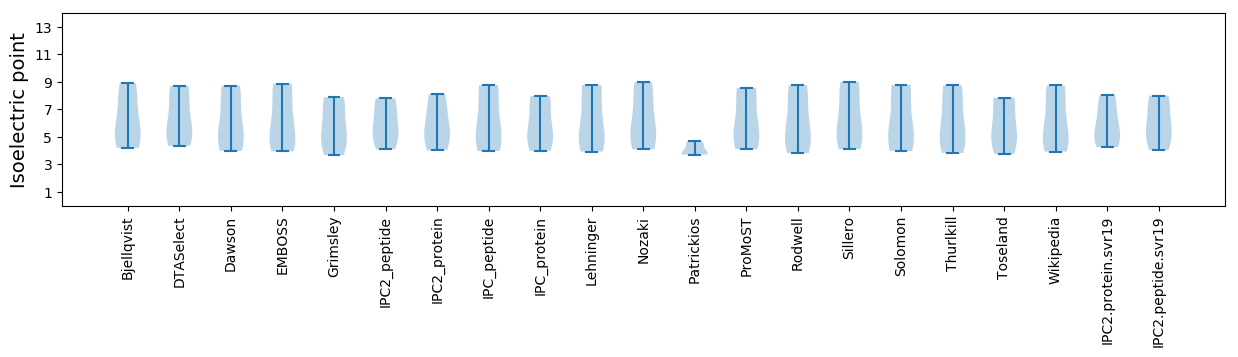
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W408|A0A4P8W408_9VIRU Uncharacterized protein OS=Capybara microvirus Cap1_SP_84 OX=2584795 PE=4 SV=1
MM1 pKa = 7.54LVTKK5 pKa = 10.51QFTDD9 pKa = 3.57EE10 pKa = 4.22SRR12 pKa = 11.84CAQLIGSHH20 pKa = 5.53NHH22 pKa = 6.92DD23 pKa = 4.45DD24 pKa = 3.64SLIYY28 pKa = 10.72VPSKK32 pKa = 10.51FMKK35 pKa = 10.48GDD37 pKa = 3.6VEE39 pKa = 4.65SVSLNSDD46 pKa = 2.83IVGYY50 pKa = 10.92ASDD53 pKa = 4.81FSLEE57 pKa = 4.06SLQAAGVDD65 pKa = 3.74PGSIAPNLSTDD76 pKa = 3.4RR77 pKa = 11.84TRR79 pKa = 11.84PNMDD83 pKa = 3.44KK84 pKa = 10.85EE85 pKa = 4.25VDD87 pKa = 3.46LSSFAEE93 pKa = 4.34SLSSLQVDD101 pKa = 3.96DD102 pKa = 5.7DD103 pKa = 4.33LPEE106 pKa = 3.84
MM1 pKa = 7.54LVTKK5 pKa = 10.51QFTDD9 pKa = 3.57EE10 pKa = 4.22SRR12 pKa = 11.84CAQLIGSHH20 pKa = 5.53NHH22 pKa = 6.92DD23 pKa = 4.45DD24 pKa = 3.64SLIYY28 pKa = 10.72VPSKK32 pKa = 10.51FMKK35 pKa = 10.48GDD37 pKa = 3.6VEE39 pKa = 4.65SVSLNSDD46 pKa = 2.83IVGYY50 pKa = 10.92ASDD53 pKa = 4.81FSLEE57 pKa = 4.06SLQAAGVDD65 pKa = 3.74PGSIAPNLSTDD76 pKa = 3.4RR77 pKa = 11.84TRR79 pKa = 11.84PNMDD83 pKa = 3.44KK84 pKa = 10.85EE85 pKa = 4.25VDD87 pKa = 3.46LSSFAEE93 pKa = 4.34SLSSLQVDD101 pKa = 3.96DD102 pKa = 5.7DD103 pKa = 4.33LPEE106 pKa = 3.84
Molecular weight: 11.5 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W4B0|A0A4P8W4B0_9VIRU Uncharacterized protein OS=Capybara microvirus Cap1_SP_84 OX=2584795 PE=4 SV=1
MM1 pKa = 7.11QATITFSNRR10 pKa = 11.84KK11 pKa = 9.52KK12 pKa = 10.86EE13 pKa = 4.05DD14 pKa = 3.65FNPLFDD20 pKa = 4.18RR21 pKa = 11.84EE22 pKa = 4.61KK23 pKa = 9.84ITIQNRR29 pKa = 11.84RR30 pKa = 11.84LLKK33 pKa = 10.35FQAAVQGGYY42 pKa = 6.58EE43 pKa = 3.58TRR45 pKa = 11.84MYY47 pKa = 10.98YY48 pKa = 10.32EE49 pKa = 4.35YY50 pKa = 10.99LNCIDD55 pKa = 4.02KK56 pKa = 11.14GGFVAFVTLTYY67 pKa = 10.92NNKK70 pKa = 9.91ALYY73 pKa = 9.57HH74 pKa = 6.27FYY76 pKa = 10.97GLPVPDD82 pKa = 4.16SRR84 pKa = 11.84NLRR87 pKa = 11.84YY88 pKa = 9.62LLHH91 pKa = 7.34DD92 pKa = 3.47SYY94 pKa = 11.16FAHH97 pKa = 6.51EE98 pKa = 4.62CARR101 pKa = 11.84SGYY104 pKa = 7.86NFKK107 pKa = 11.17YY108 pKa = 10.4FITTEE113 pKa = 3.93LGEE116 pKa = 4.56GKK118 pKa = 10.31GKK120 pKa = 10.51RR121 pKa = 11.84GFANNPHH128 pKa = 5.38YY129 pKa = 10.8HH130 pKa = 6.46CLLYY134 pKa = 8.03FTPSTPEE141 pKa = 3.52YY142 pKa = 10.25NRR144 pKa = 11.84FFITRR149 pKa = 11.84YY150 pKa = 8.92VDD152 pKa = 4.57LIKK155 pKa = 10.69KK156 pKa = 8.35YY157 pKa = 8.1WQGDD161 pKa = 3.37NGEE164 pKa = 4.56DD165 pKa = 2.88WNAAKK170 pKa = 10.24FGHH173 pKa = 5.99VEE175 pKa = 3.77PGDD178 pKa = 3.33NYY180 pKa = 11.32GIVNSSSAFSYY191 pKa = 9.03VSKK194 pKa = 10.99YY195 pKa = 6.57VTKK198 pKa = 10.67SSTDD202 pKa = 2.77IHH204 pKa = 6.94RR205 pKa = 11.84EE206 pKa = 3.74RR207 pKa = 11.84IVAARR212 pKa = 11.84SKK214 pKa = 10.21EE215 pKa = 4.02YY216 pKa = 9.41WSKK219 pKa = 11.0ILDD222 pKa = 3.9PCSSEE227 pKa = 5.02FKK229 pKa = 11.16VLDD232 pKa = 3.43IKK234 pKa = 11.49LNDD237 pKa = 4.86LIKK240 pKa = 10.82ADD242 pKa = 3.28INKK245 pKa = 8.97YY246 pKa = 10.15RR247 pKa = 11.84NLFGTRR253 pKa = 11.84VRR255 pKa = 11.84CSHH258 pKa = 6.38GFGQIDD264 pKa = 3.65EE265 pKa = 4.45TLLTFIHH272 pKa = 7.09DD273 pKa = 3.87KK274 pKa = 10.74PHH276 pKa = 7.12IKK278 pKa = 9.17LTSNGKK284 pKa = 8.25LTYY287 pKa = 10.18KK288 pKa = 10.45PLPLYY293 pKa = 10.4NYY295 pKa = 8.84RR296 pKa = 11.84KK297 pKa = 9.79LYY299 pKa = 9.99MYY301 pKa = 10.91VDD303 pKa = 3.47EE304 pKa = 4.65NKK306 pKa = 10.25CYY308 pKa = 10.52KK309 pKa = 10.54LNEE312 pKa = 4.04AGIEE316 pKa = 4.18LKK318 pKa = 10.62CRR320 pKa = 11.84QVSSALKK327 pKa = 10.83DD328 pKa = 3.47MADD331 pKa = 3.57NAQLICRR338 pKa = 11.84NTLSCCEE345 pKa = 4.12VFSLLPSPLDD355 pKa = 3.34LSTYY359 pKa = 9.78IQQCNCKK366 pKa = 10.09LGEE369 pKa = 4.17SPLFHH374 pKa = 7.86DD375 pKa = 4.07YY376 pKa = 11.6ALYY379 pKa = 9.59TLLYY383 pKa = 8.8KK384 pKa = 10.87DD385 pKa = 3.92RR386 pKa = 11.84MYY388 pKa = 10.78PSWPSGEE395 pKa = 4.73FFDD398 pKa = 5.79EE399 pKa = 4.32PDD401 pKa = 4.15IDD403 pKa = 3.79PLSYY407 pKa = 10.03IRR409 pKa = 11.84QVSSRR414 pKa = 11.84VDD416 pKa = 3.0KK417 pKa = 11.23NSYY420 pKa = 9.18FFCSDD425 pKa = 3.63CAHH428 pKa = 7.26LLPYY432 pKa = 9.89KK433 pKa = 8.98YY434 pKa = 9.96HH435 pKa = 5.95SRR437 pKa = 11.84FRR439 pKa = 11.84FNLPFFSAIDD449 pKa = 3.68SVLSHH454 pKa = 6.21HH455 pKa = 7.44TIQTDD460 pKa = 3.29EE461 pKa = 4.82KK462 pKa = 9.47LTQEE466 pKa = 3.76HH467 pKa = 6.28EE468 pKa = 3.78LRR470 pKa = 11.84KK471 pKa = 9.89RR472 pKa = 11.84IRR474 pKa = 11.84NSVLAQRR481 pKa = 11.84KK482 pKa = 8.05PFLFHH487 pKa = 7.16SYY489 pKa = 10.38KK490 pKa = 10.83
MM1 pKa = 7.11QATITFSNRR10 pKa = 11.84KK11 pKa = 9.52KK12 pKa = 10.86EE13 pKa = 4.05DD14 pKa = 3.65FNPLFDD20 pKa = 4.18RR21 pKa = 11.84EE22 pKa = 4.61KK23 pKa = 9.84ITIQNRR29 pKa = 11.84RR30 pKa = 11.84LLKK33 pKa = 10.35FQAAVQGGYY42 pKa = 6.58EE43 pKa = 3.58TRR45 pKa = 11.84MYY47 pKa = 10.98YY48 pKa = 10.32EE49 pKa = 4.35YY50 pKa = 10.99LNCIDD55 pKa = 4.02KK56 pKa = 11.14GGFVAFVTLTYY67 pKa = 10.92NNKK70 pKa = 9.91ALYY73 pKa = 9.57HH74 pKa = 6.27FYY76 pKa = 10.97GLPVPDD82 pKa = 4.16SRR84 pKa = 11.84NLRR87 pKa = 11.84YY88 pKa = 9.62LLHH91 pKa = 7.34DD92 pKa = 3.47SYY94 pKa = 11.16FAHH97 pKa = 6.51EE98 pKa = 4.62CARR101 pKa = 11.84SGYY104 pKa = 7.86NFKK107 pKa = 11.17YY108 pKa = 10.4FITTEE113 pKa = 3.93LGEE116 pKa = 4.56GKK118 pKa = 10.31GKK120 pKa = 10.51RR121 pKa = 11.84GFANNPHH128 pKa = 5.38YY129 pKa = 10.8HH130 pKa = 6.46CLLYY134 pKa = 8.03FTPSTPEE141 pKa = 3.52YY142 pKa = 10.25NRR144 pKa = 11.84FFITRR149 pKa = 11.84YY150 pKa = 8.92VDD152 pKa = 4.57LIKK155 pKa = 10.69KK156 pKa = 8.35YY157 pKa = 8.1WQGDD161 pKa = 3.37NGEE164 pKa = 4.56DD165 pKa = 2.88WNAAKK170 pKa = 10.24FGHH173 pKa = 5.99VEE175 pKa = 3.77PGDD178 pKa = 3.33NYY180 pKa = 11.32GIVNSSSAFSYY191 pKa = 9.03VSKK194 pKa = 10.99YY195 pKa = 6.57VTKK198 pKa = 10.67SSTDD202 pKa = 2.77IHH204 pKa = 6.94RR205 pKa = 11.84EE206 pKa = 3.74RR207 pKa = 11.84IVAARR212 pKa = 11.84SKK214 pKa = 10.21EE215 pKa = 4.02YY216 pKa = 9.41WSKK219 pKa = 11.0ILDD222 pKa = 3.9PCSSEE227 pKa = 5.02FKK229 pKa = 11.16VLDD232 pKa = 3.43IKK234 pKa = 11.49LNDD237 pKa = 4.86LIKK240 pKa = 10.82ADD242 pKa = 3.28INKK245 pKa = 8.97YY246 pKa = 10.15RR247 pKa = 11.84NLFGTRR253 pKa = 11.84VRR255 pKa = 11.84CSHH258 pKa = 6.38GFGQIDD264 pKa = 3.65EE265 pKa = 4.45TLLTFIHH272 pKa = 7.09DD273 pKa = 3.87KK274 pKa = 10.74PHH276 pKa = 7.12IKK278 pKa = 9.17LTSNGKK284 pKa = 8.25LTYY287 pKa = 10.18KK288 pKa = 10.45PLPLYY293 pKa = 10.4NYY295 pKa = 8.84RR296 pKa = 11.84KK297 pKa = 9.79LYY299 pKa = 9.99MYY301 pKa = 10.91VDD303 pKa = 3.47EE304 pKa = 4.65NKK306 pKa = 10.25CYY308 pKa = 10.52KK309 pKa = 10.54LNEE312 pKa = 4.04AGIEE316 pKa = 4.18LKK318 pKa = 10.62CRR320 pKa = 11.84QVSSALKK327 pKa = 10.83DD328 pKa = 3.47MADD331 pKa = 3.57NAQLICRR338 pKa = 11.84NTLSCCEE345 pKa = 4.12VFSLLPSPLDD355 pKa = 3.34LSTYY359 pKa = 9.78IQQCNCKK366 pKa = 10.09LGEE369 pKa = 4.17SPLFHH374 pKa = 7.86DD375 pKa = 4.07YY376 pKa = 11.6ALYY379 pKa = 9.59TLLYY383 pKa = 8.8KK384 pKa = 10.87DD385 pKa = 3.92RR386 pKa = 11.84MYY388 pKa = 10.78PSWPSGEE395 pKa = 4.73FFDD398 pKa = 5.79EE399 pKa = 4.32PDD401 pKa = 4.15IDD403 pKa = 3.79PLSYY407 pKa = 10.03IRR409 pKa = 11.84QVSSRR414 pKa = 11.84VDD416 pKa = 3.0KK417 pKa = 11.23NSYY420 pKa = 9.18FFCSDD425 pKa = 3.63CAHH428 pKa = 7.26LLPYY432 pKa = 9.89KK433 pKa = 8.98YY434 pKa = 9.96HH435 pKa = 5.95SRR437 pKa = 11.84FRR439 pKa = 11.84FNLPFFSAIDD449 pKa = 3.68SVLSHH454 pKa = 6.21HH455 pKa = 7.44TIQTDD460 pKa = 3.29EE461 pKa = 4.82KK462 pKa = 9.47LTQEE466 pKa = 3.76HH467 pKa = 6.28EE468 pKa = 3.78LRR470 pKa = 11.84KK471 pKa = 9.89RR472 pKa = 11.84IRR474 pKa = 11.84NSVLAQRR481 pKa = 11.84KK482 pKa = 8.05PFLFHH487 pKa = 7.16SYY489 pKa = 10.38KK490 pKa = 10.83
Molecular weight: 57.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1678 |
78 |
603 |
335.6 |
38.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.555 ± 1.232 | 1.192 ± 0.581 |
6.436 ± 0.543 | 5.959 ± 0.879 |
5.662 ± 0.639 | 5.304 ± 0.724 |
2.086 ± 0.52 | 4.41 ± 0.26 |
5.9 ± 0.636 | 9.178 ± 0.368 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.907 ± 0.328 | 7.032 ± 1.056 |
3.456 ± 0.508 | 4.529 ± 1.176 |
5.304 ± 0.482 | 8.522 ± 0.66 |
4.768 ± 0.601 | 5.662 ± 0.848 |
1.073 ± 0.287 | 5.066 ± 1.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
