
Daphnia pulex (Water flea)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Panarthropoda; Arthropoda; Mandibulata; Pancrustacea; Crustacea; Branchiopoda; Phyllopoda; Diplostraca; Cladocera; Anomopoda; Daphniidae; Daphnia
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
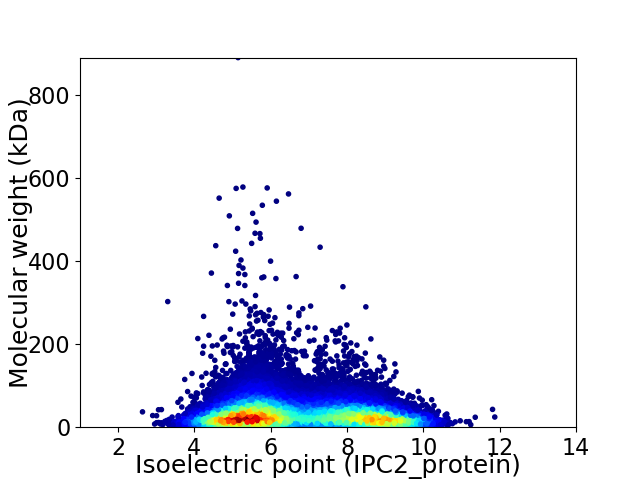
Virtual 2D-PAGE plot for 30137 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E9GEE1|E9GEE1_DAPPU PBPe domain-containing protein OS=Daphnia pulex OX=6669 GN=DAPPUDRAFT_101737 PE=4 SV=1
MM1 pKa = 7.4SSQVSFLVLMAMAVVALTGTLGSCSIIIRR30 pKa = 11.84SASAGDD36 pKa = 4.07YY37 pKa = 11.13YY38 pKa = 11.34DD39 pKa = 5.93LNQQQLVLSPEE50 pKa = 3.37IRR52 pKa = 11.84AKK54 pKa = 9.93VVRR57 pKa = 11.84PYY59 pKa = 11.51APLQNEE65 pKa = 4.1LTEE68 pKa = 4.21EE69 pKa = 3.96NLAKK73 pKa = 10.59VEE75 pKa = 4.12EE76 pKa = 4.56LTTDD80 pKa = 3.2VPTDD84 pKa = 3.46EE85 pKa = 4.7TTDD88 pKa = 3.33ATTDD92 pKa = 3.27APTNAPTDD100 pKa = 3.96TPTDD104 pKa = 3.63TPTDD108 pKa = 3.78EE109 pKa = 4.72PTDD112 pKa = 3.94KK113 pKa = 9.98PTDD116 pKa = 3.27AHH118 pKa = 6.35TDD120 pKa = 3.35APTDD124 pKa = 3.54APTNAPTDD132 pKa = 3.75APTDD136 pKa = 3.73APTDD140 pKa = 3.66APTNAPTNAPTDD152 pKa = 3.34AHH154 pKa = 6.4TDD156 pKa = 3.13APTNVPTDD164 pKa = 3.64APTDD168 pKa = 3.74APTEE172 pKa = 4.15APTNVPTDD180 pKa = 3.64APTDD184 pKa = 3.75APTDD188 pKa = 3.73APTDD192 pKa = 3.66APTNAPTDD200 pKa = 3.77APTQAPTDD208 pKa = 3.94APTQAPTDD216 pKa = 3.79APTNAPTNAPTDD228 pKa = 3.34AHH230 pKa = 6.33TDD232 pKa = 3.4APTDD236 pKa = 3.63APTQAPTDD244 pKa = 3.79APTNAPTDD252 pKa = 3.75APTDD256 pKa = 3.66APTDD260 pKa = 3.73EE261 pKa = 5.19PPNTTADD268 pKa = 3.74PTTDD272 pKa = 3.16TTTEE276 pKa = 4.02TSTDD280 pKa = 3.22TSTDD284 pKa = 2.76ATTATTVSTTTTTTTTTTAATVSTTTPVTEE314 pKa = 4.39ASTTTSVTTAPFSSSTATISSTSARR339 pKa = 11.84TTTSAPSSGNIFTRR353 pKa = 11.84VQAHH357 pKa = 6.41LEE359 pKa = 4.6FIQSHH364 pKa = 5.24TKK366 pKa = 9.67VSSPADD372 pKa = 3.42CSSTTNGTEE381 pKa = 3.9PTTTPGPTTSTTQKK395 pKa = 8.96ITTTSTRR402 pKa = 11.84KK403 pKa = 6.25TTTSTTTASTTNQSVTFSCAGKK425 pKa = 10.77ANGNNPNPASSCSNTFYY442 pKa = 11.25ACSNGNACLFNCPSNLVYY460 pKa = 10.5RR461 pKa = 11.84EE462 pKa = 4.1EE463 pKa = 5.09IGVCDD468 pKa = 3.9YY469 pKa = 10.42PSNVAGCYY477 pKa = 9.32
MM1 pKa = 7.4SSQVSFLVLMAMAVVALTGTLGSCSIIIRR30 pKa = 11.84SASAGDD36 pKa = 4.07YY37 pKa = 11.13YY38 pKa = 11.34DD39 pKa = 5.93LNQQQLVLSPEE50 pKa = 3.37IRR52 pKa = 11.84AKK54 pKa = 9.93VVRR57 pKa = 11.84PYY59 pKa = 11.51APLQNEE65 pKa = 4.1LTEE68 pKa = 4.21EE69 pKa = 3.96NLAKK73 pKa = 10.59VEE75 pKa = 4.12EE76 pKa = 4.56LTTDD80 pKa = 3.2VPTDD84 pKa = 3.46EE85 pKa = 4.7TTDD88 pKa = 3.33ATTDD92 pKa = 3.27APTNAPTDD100 pKa = 3.96TPTDD104 pKa = 3.63TPTDD108 pKa = 3.78EE109 pKa = 4.72PTDD112 pKa = 3.94KK113 pKa = 9.98PTDD116 pKa = 3.27AHH118 pKa = 6.35TDD120 pKa = 3.35APTDD124 pKa = 3.54APTNAPTDD132 pKa = 3.75APTDD136 pKa = 3.73APTDD140 pKa = 3.66APTNAPTNAPTDD152 pKa = 3.34AHH154 pKa = 6.4TDD156 pKa = 3.13APTNVPTDD164 pKa = 3.64APTDD168 pKa = 3.74APTEE172 pKa = 4.15APTNVPTDD180 pKa = 3.64APTDD184 pKa = 3.75APTDD188 pKa = 3.73APTDD192 pKa = 3.66APTNAPTDD200 pKa = 3.77APTQAPTDD208 pKa = 3.94APTQAPTDD216 pKa = 3.79APTNAPTNAPTDD228 pKa = 3.34AHH230 pKa = 6.33TDD232 pKa = 3.4APTDD236 pKa = 3.63APTQAPTDD244 pKa = 3.79APTNAPTDD252 pKa = 3.75APTDD256 pKa = 3.66APTDD260 pKa = 3.73EE261 pKa = 5.19PPNTTADD268 pKa = 3.74PTTDD272 pKa = 3.16TTTEE276 pKa = 4.02TSTDD280 pKa = 3.22TSTDD284 pKa = 2.76ATTATTVSTTTTTTTTTTAATVSTTTPVTEE314 pKa = 4.39ASTTTSVTTAPFSSSTATISSTSARR339 pKa = 11.84TTTSAPSSGNIFTRR353 pKa = 11.84VQAHH357 pKa = 6.41LEE359 pKa = 4.6FIQSHH364 pKa = 5.24TKK366 pKa = 9.67VSSPADD372 pKa = 3.42CSSTTNGTEE381 pKa = 3.9PTTTPGPTTSTTQKK395 pKa = 8.96ITTTSTRR402 pKa = 11.84KK403 pKa = 6.25TTTSTTTASTTNQSVTFSCAGKK425 pKa = 10.77ANGNNPNPASSCSNTFYY442 pKa = 11.25ACSNGNACLFNCPSNLVYY460 pKa = 10.5RR461 pKa = 11.84EE462 pKa = 4.1EE463 pKa = 5.09IGVCDD468 pKa = 3.9YY469 pKa = 10.42PSNVAGCYY477 pKa = 9.32
Molecular weight: 48.59 kDa
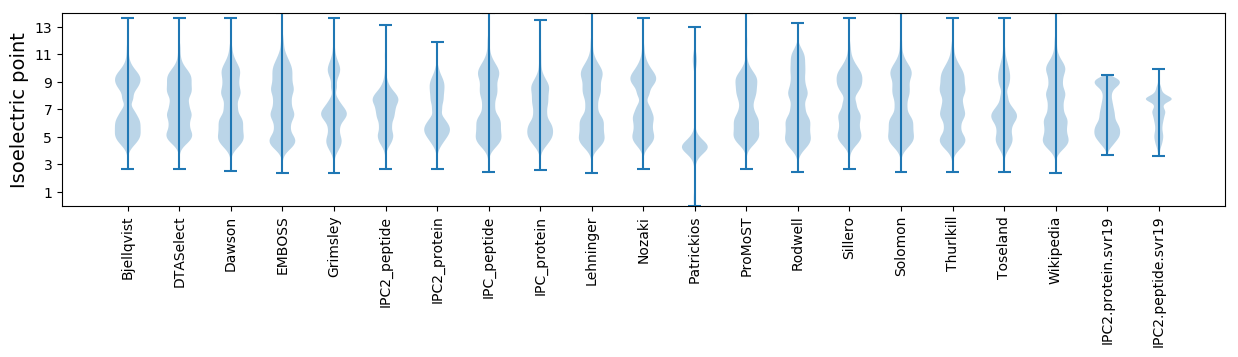
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E9I6K4|E9I6K4_DAPPU Uncharacterized protein OS=Daphnia pulex OX=6669 GN=DAPPUDRAFT_124402 PE=4 SV=1
AA1 pKa = 7.54QAQAQASPRR10 pKa = 11.84FAKK13 pKa = 9.93VRR15 pKa = 11.84QGSPRR20 pKa = 11.84FAKK23 pKa = 9.89VRR25 pKa = 11.84QGSPRR30 pKa = 11.84FAEE33 pKa = 4.29VRR35 pKa = 11.84RR36 pKa = 11.84GSPRR40 pKa = 11.84FAEE43 pKa = 4.49VRR45 pKa = 11.84QGSPRR50 pKa = 11.84FAKK53 pKa = 9.89VRR55 pKa = 11.84QGSPRR60 pKa = 11.84FAKK63 pKa = 9.89VRR65 pKa = 11.84QGSPRR70 pKa = 11.84FAKK73 pKa = 9.89VRR75 pKa = 11.84QGSPRR80 pKa = 11.84FAKK83 pKa = 9.89VRR85 pKa = 11.84QGSPRR90 pKa = 11.84FAKK93 pKa = 9.89VRR95 pKa = 11.84QGSPRR100 pKa = 11.84FAKK103 pKa = 9.89VRR105 pKa = 11.84QGSPRR110 pKa = 11.84FAKK113 pKa = 9.89VRR115 pKa = 11.84QGSPRR120 pKa = 11.84FAKK123 pKa = 9.89VRR125 pKa = 11.84QGSPRR130 pKa = 11.84FAKK133 pKa = 9.89VRR135 pKa = 11.84QGSPRR140 pKa = 11.84FAKK143 pKa = 9.89VRR145 pKa = 11.84QGSPRR150 pKa = 11.84FAKK153 pKa = 9.89VRR155 pKa = 11.84QGSPRR160 pKa = 11.84FAKK163 pKa = 9.89VRR165 pKa = 11.84QGSPRR170 pKa = 11.84FAKK173 pKa = 9.89VRR175 pKa = 11.84QGSPRR180 pKa = 11.84FAKK183 pKa = 9.89VRR185 pKa = 11.84QGSPRR190 pKa = 11.84FAKK193 pKa = 9.89VRR195 pKa = 11.84QGSPRR200 pKa = 11.84FAKK203 pKa = 9.89VRR205 pKa = 11.84QGSPRR210 pKa = 11.84FAKK213 pKa = 10.51
AA1 pKa = 7.54QAQAQASPRR10 pKa = 11.84FAKK13 pKa = 9.93VRR15 pKa = 11.84QGSPRR20 pKa = 11.84FAKK23 pKa = 9.89VRR25 pKa = 11.84QGSPRR30 pKa = 11.84FAEE33 pKa = 4.29VRR35 pKa = 11.84RR36 pKa = 11.84GSPRR40 pKa = 11.84FAEE43 pKa = 4.49VRR45 pKa = 11.84QGSPRR50 pKa = 11.84FAKK53 pKa = 9.89VRR55 pKa = 11.84QGSPRR60 pKa = 11.84FAKK63 pKa = 9.89VRR65 pKa = 11.84QGSPRR70 pKa = 11.84FAKK73 pKa = 9.89VRR75 pKa = 11.84QGSPRR80 pKa = 11.84FAKK83 pKa = 9.89VRR85 pKa = 11.84QGSPRR90 pKa = 11.84FAKK93 pKa = 9.89VRR95 pKa = 11.84QGSPRR100 pKa = 11.84FAKK103 pKa = 9.89VRR105 pKa = 11.84QGSPRR110 pKa = 11.84FAKK113 pKa = 9.89VRR115 pKa = 11.84QGSPRR120 pKa = 11.84FAKK123 pKa = 9.89VRR125 pKa = 11.84QGSPRR130 pKa = 11.84FAKK133 pKa = 9.89VRR135 pKa = 11.84QGSPRR140 pKa = 11.84FAKK143 pKa = 9.89VRR145 pKa = 11.84QGSPRR150 pKa = 11.84FAKK153 pKa = 9.89VRR155 pKa = 11.84QGSPRR160 pKa = 11.84FAKK163 pKa = 9.89VRR165 pKa = 11.84QGSPRR170 pKa = 11.84FAKK173 pKa = 9.89VRR175 pKa = 11.84QGSPRR180 pKa = 11.84FAKK183 pKa = 9.89VRR185 pKa = 11.84QGSPRR190 pKa = 11.84FAKK193 pKa = 9.89VRR195 pKa = 11.84QGSPRR200 pKa = 11.84FAKK203 pKa = 9.89VRR205 pKa = 11.84QGSPRR210 pKa = 11.84FAKK213 pKa = 10.51
Molecular weight: 23.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
9947364 |
20 |
7776 |
330.1 |
36.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.936 ± 0.018 | 1.928 ± 0.015 |
5.296 ± 0.012 | 6.405 ± 0.02 |
3.962 ± 0.013 | 5.692 ± 0.019 |
2.445 ± 0.008 | 5.278 ± 0.011 |
5.719 ± 0.018 | 9.161 ± 0.02 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.261 ± 0.007 | 4.655 ± 0.01 |
5.431 ± 0.018 | 4.44 ± 0.012 |
5.514 ± 0.014 | 8.381 ± 0.021 |
5.934 ± 0.02 | 6.41 ± 0.017 |
1.197 ± 0.006 | 2.952 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
