
Sus scrofa papillomavirus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; unclassified Papillomaviridae; non-primate mammal papillomaviruses
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
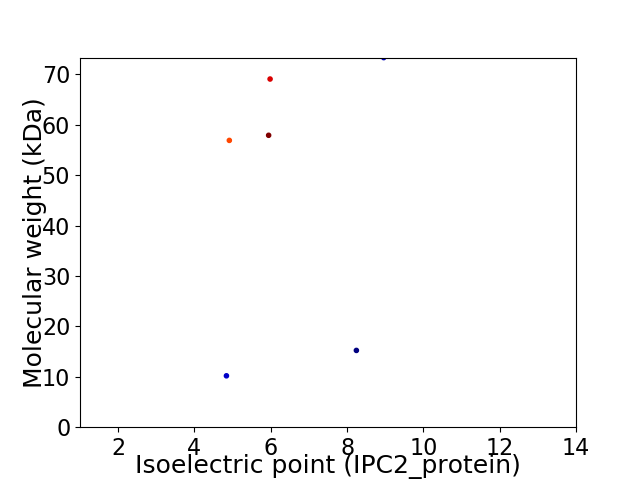
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A223FQV1|A0A223FQV1_9PAPI Regulatory protein E2 OS=Sus scrofa papillomavirus 2 OX=2025338 GN=E2 PE=3 SV=1
MM1 pKa = 7.11MGVEE5 pKa = 4.0PTLRR9 pKa = 11.84DD10 pKa = 3.5IVLMDD15 pKa = 4.54LPCPLHH21 pKa = 6.48LGCEE25 pKa = 4.04EE26 pKa = 4.1RR27 pKa = 11.84LEE29 pKa = 5.23DD30 pKa = 4.51MMLPEE35 pKa = 5.74DD36 pKa = 4.95DD37 pKa = 4.54EE38 pKa = 6.28ASEE41 pKa = 4.41APYY44 pKa = 10.15KK45 pKa = 11.06VSVLCGLCRR54 pKa = 11.84KK55 pKa = 8.76PLKK58 pKa = 10.57FLVICSGEE66 pKa = 3.99NIQRR70 pKa = 11.84LEE72 pKa = 4.0DD73 pKa = 3.07VAARR77 pKa = 11.84VKK79 pKa = 9.92YY80 pKa = 10.63LCRR83 pKa = 11.84CCSARR88 pKa = 11.84FDD90 pKa = 3.59
MM1 pKa = 7.11MGVEE5 pKa = 4.0PTLRR9 pKa = 11.84DD10 pKa = 3.5IVLMDD15 pKa = 4.54LPCPLHH21 pKa = 6.48LGCEE25 pKa = 4.04EE26 pKa = 4.1RR27 pKa = 11.84LEE29 pKa = 5.23DD30 pKa = 4.51MMLPEE35 pKa = 5.74DD36 pKa = 4.95DD37 pKa = 4.54EE38 pKa = 6.28ASEE41 pKa = 4.41APYY44 pKa = 10.15KK45 pKa = 11.06VSVLCGLCRR54 pKa = 11.84KK55 pKa = 8.76PLKK58 pKa = 10.57FLVICSGEE66 pKa = 3.99NIQRR70 pKa = 11.84LEE72 pKa = 4.0DD73 pKa = 3.07VAARR77 pKa = 11.84VKK79 pKa = 9.92YY80 pKa = 10.63LCRR83 pKa = 11.84CCSARR88 pKa = 11.84FDD90 pKa = 3.59
Molecular weight: 10.19 kDa
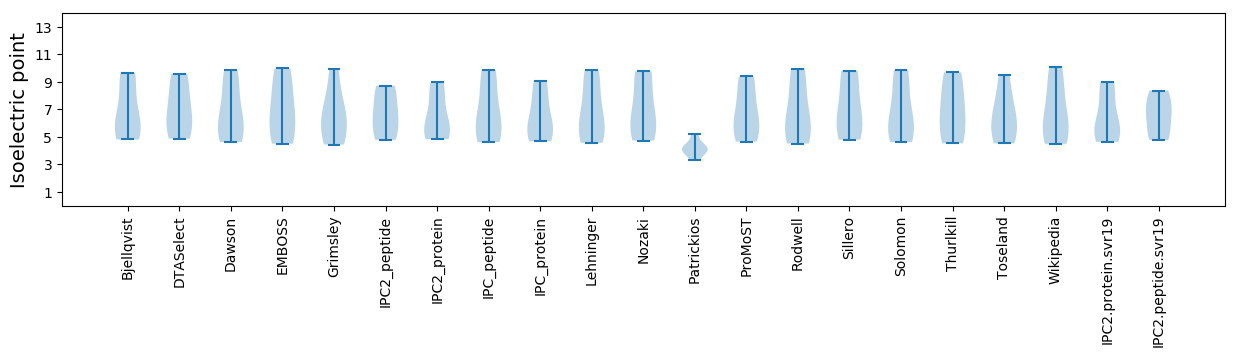
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A223FQV2|A0A223FQV2_9PAPI Major capsid protein L1 OS=Sus scrofa papillomavirus 2 OX=2025338 GN=L1 PE=3 SV=1
MM1 pKa = 7.43EE2 pKa = 6.17RR3 pKa = 11.84ICHH6 pKa = 6.31RR7 pKa = 11.84LNVVQEE13 pKa = 4.3EE14 pKa = 4.35LLSLYY19 pKa = 7.87EE20 pKa = 4.02TAQTSLQSQIRR31 pKa = 11.84HH32 pKa = 5.36WDD34 pKa = 3.5LVRR37 pKa = 11.84KK38 pKa = 9.71EE39 pKa = 4.35MIFFYY44 pKa = 10.37AARR47 pKa = 11.84QSGVDD52 pKa = 3.3SIGFQKK58 pKa = 10.43VPPLQVSQAKK68 pKa = 9.82AKK70 pKa = 10.07EE71 pKa = 4.19AIEE74 pKa = 4.0MQMILQSLGEE84 pKa = 4.02SDD86 pKa = 4.37FAQEE90 pKa = 3.72QWTLSDD96 pKa = 3.3TSRR99 pKa = 11.84EE100 pKa = 3.9LWLCPPQGCFKK111 pKa = 10.76KK112 pKa = 9.92HH113 pKa = 5.46GSSVDD118 pKa = 2.87VWYY121 pKa = 10.33DD122 pKa = 3.74EE123 pKa = 4.31NPEE126 pKa = 3.85NANRR130 pKa = 11.84YY131 pKa = 8.1VSWGSIYY138 pKa = 10.67HH139 pKa = 6.86LDD141 pKa = 3.75GNEE144 pKa = 3.61RR145 pKa = 11.84WRR147 pKa = 11.84KK148 pKa = 7.47TEE150 pKa = 3.79GLCDD154 pKa = 3.58HH155 pKa = 7.01TGLYY159 pKa = 10.17YY160 pKa = 10.47VDD162 pKa = 3.25EE163 pKa = 4.6SGYY166 pKa = 9.6RR167 pKa = 11.84RR168 pKa = 11.84YY169 pKa = 10.25YY170 pKa = 10.3IKK172 pKa = 10.71FSVDD176 pKa = 2.64AKK178 pKa = 10.31RR179 pKa = 11.84YY180 pKa = 8.49SATGQWEE187 pKa = 4.64VKK189 pKa = 9.94FKK191 pKa = 11.2NQIFSPILPVTSTVPATQCGGPAPEE216 pKa = 4.17SCGRR220 pKa = 11.84QTPEE224 pKa = 3.61QQRR227 pKa = 11.84IASPPGGHH235 pKa = 6.07GVSPPASRR243 pKa = 11.84CRR245 pKa = 11.84GASPGTDD252 pKa = 2.64RR253 pKa = 11.84ADD255 pKa = 3.54TPPPKK260 pKa = 9.89RR261 pKa = 11.84PRR263 pKa = 11.84DD264 pKa = 3.65TRR266 pKa = 11.84PTEE269 pKa = 3.76TSKK272 pKa = 10.73GRR274 pKa = 11.84RR275 pKa = 11.84RR276 pKa = 11.84RR277 pKa = 11.84SIRR280 pKa = 11.84PGLGPRR286 pKa = 11.84TSIVATGPVWVPPIIRR302 pKa = 11.84PPRR305 pKa = 11.84FPGSGSPWAARR316 pKa = 11.84WYY318 pKa = 10.75AKK320 pKa = 10.45FEE322 pKa = 3.84EE323 pKa = 4.67RR324 pKa = 11.84QRR326 pKa = 11.84QQQQQQQQQQQAPIRR341 pKa = 11.84FPGPADD347 pKa = 3.46TGDD350 pKa = 3.64SSGSRR355 pKa = 11.84VSRR358 pKa = 11.84DD359 pKa = 3.35PGHH362 pKa = 6.6SSRR365 pKa = 11.84DD366 pKa = 3.4PSDD369 pKa = 3.9PGDD372 pKa = 3.53TRR374 pKa = 11.84EE375 pKa = 4.86PGDD378 pKa = 3.61PCDD381 pKa = 4.01PSEE384 pKa = 4.64EE385 pKa = 4.5GPATEE390 pKa = 4.84SDD392 pKa = 3.75SGSDD396 pKa = 3.04SDD398 pKa = 4.05PNTRR402 pKa = 11.84RR403 pKa = 11.84EE404 pKa = 4.28SPPGPWDD411 pKa = 3.49TRR413 pKa = 11.84PTGPQPPRR421 pKa = 11.84TPEE424 pKa = 3.48EE425 pKa = 3.75AAAAAAATVTPPGAKK440 pKa = 9.03EE441 pKa = 3.24KK442 pKa = 10.42AYY444 pKa = 10.8RR445 pKa = 11.84EE446 pKa = 4.12GRR448 pKa = 11.84GLRR451 pKa = 11.84RR452 pKa = 11.84QRR454 pKa = 11.84PPTPPEE460 pKa = 3.98CYY462 pKa = 9.4RR463 pKa = 11.84RR464 pKa = 11.84VPFTRR469 pKa = 11.84QQSTKK474 pKa = 10.41SRR476 pKa = 11.84NTTLVSPKK484 pKa = 8.64QTPVKK489 pKa = 10.11QGPSSPLAASTPAAATRR506 pKa = 11.84SRR508 pKa = 11.84TTRR511 pKa = 11.84RR512 pKa = 11.84NRR514 pKa = 11.84SRR516 pKa = 11.84SPTNPVASQPVGIAGRR532 pKa = 11.84TRR534 pKa = 11.84RR535 pKa = 11.84SRR537 pKa = 11.84YY538 pKa = 8.21PVEE541 pKa = 4.27EE542 pKa = 4.34EE543 pKa = 4.34SCGGDD548 pKa = 3.13PGGTLQSLRR557 pKa = 11.84DD558 pKa = 3.58PDD560 pKa = 3.55NQRR563 pKa = 11.84EE564 pKa = 4.05QRR566 pKa = 11.84PGQSLPVLGSPAVVLLKK583 pKa = 11.04GGTNQLKK590 pKa = 8.58CCRR593 pKa = 11.84YY594 pKa = 9.45RR595 pKa = 11.84FRR597 pKa = 11.84TRR599 pKa = 11.84HH600 pKa = 3.5QHH602 pKa = 6.71LFLHH606 pKa = 7.08CSSTWYY612 pKa = 9.56WSDD615 pKa = 2.8GNRR618 pKa = 11.84IGRR621 pKa = 11.84SRR623 pKa = 11.84MFLTFRR629 pKa = 11.84DD630 pKa = 3.97PEE632 pKa = 4.05QRR634 pKa = 11.84DD635 pKa = 3.46LFLQTVSIPRR645 pKa = 11.84DD646 pKa = 3.19VDD648 pKa = 3.31IVTGTLFSLL657 pKa = 4.44
MM1 pKa = 7.43EE2 pKa = 6.17RR3 pKa = 11.84ICHH6 pKa = 6.31RR7 pKa = 11.84LNVVQEE13 pKa = 4.3EE14 pKa = 4.35LLSLYY19 pKa = 7.87EE20 pKa = 4.02TAQTSLQSQIRR31 pKa = 11.84HH32 pKa = 5.36WDD34 pKa = 3.5LVRR37 pKa = 11.84KK38 pKa = 9.71EE39 pKa = 4.35MIFFYY44 pKa = 10.37AARR47 pKa = 11.84QSGVDD52 pKa = 3.3SIGFQKK58 pKa = 10.43VPPLQVSQAKK68 pKa = 9.82AKK70 pKa = 10.07EE71 pKa = 4.19AIEE74 pKa = 4.0MQMILQSLGEE84 pKa = 4.02SDD86 pKa = 4.37FAQEE90 pKa = 3.72QWTLSDD96 pKa = 3.3TSRR99 pKa = 11.84EE100 pKa = 3.9LWLCPPQGCFKK111 pKa = 10.76KK112 pKa = 9.92HH113 pKa = 5.46GSSVDD118 pKa = 2.87VWYY121 pKa = 10.33DD122 pKa = 3.74EE123 pKa = 4.31NPEE126 pKa = 3.85NANRR130 pKa = 11.84YY131 pKa = 8.1VSWGSIYY138 pKa = 10.67HH139 pKa = 6.86LDD141 pKa = 3.75GNEE144 pKa = 3.61RR145 pKa = 11.84WRR147 pKa = 11.84KK148 pKa = 7.47TEE150 pKa = 3.79GLCDD154 pKa = 3.58HH155 pKa = 7.01TGLYY159 pKa = 10.17YY160 pKa = 10.47VDD162 pKa = 3.25EE163 pKa = 4.6SGYY166 pKa = 9.6RR167 pKa = 11.84RR168 pKa = 11.84YY169 pKa = 10.25YY170 pKa = 10.3IKK172 pKa = 10.71FSVDD176 pKa = 2.64AKK178 pKa = 10.31RR179 pKa = 11.84YY180 pKa = 8.49SATGQWEE187 pKa = 4.64VKK189 pKa = 9.94FKK191 pKa = 11.2NQIFSPILPVTSTVPATQCGGPAPEE216 pKa = 4.17SCGRR220 pKa = 11.84QTPEE224 pKa = 3.61QQRR227 pKa = 11.84IASPPGGHH235 pKa = 6.07GVSPPASRR243 pKa = 11.84CRR245 pKa = 11.84GASPGTDD252 pKa = 2.64RR253 pKa = 11.84ADD255 pKa = 3.54TPPPKK260 pKa = 9.89RR261 pKa = 11.84PRR263 pKa = 11.84DD264 pKa = 3.65TRR266 pKa = 11.84PTEE269 pKa = 3.76TSKK272 pKa = 10.73GRR274 pKa = 11.84RR275 pKa = 11.84RR276 pKa = 11.84RR277 pKa = 11.84SIRR280 pKa = 11.84PGLGPRR286 pKa = 11.84TSIVATGPVWVPPIIRR302 pKa = 11.84PPRR305 pKa = 11.84FPGSGSPWAARR316 pKa = 11.84WYY318 pKa = 10.75AKK320 pKa = 10.45FEE322 pKa = 3.84EE323 pKa = 4.67RR324 pKa = 11.84QRR326 pKa = 11.84QQQQQQQQQQQAPIRR341 pKa = 11.84FPGPADD347 pKa = 3.46TGDD350 pKa = 3.64SSGSRR355 pKa = 11.84VSRR358 pKa = 11.84DD359 pKa = 3.35PGHH362 pKa = 6.6SSRR365 pKa = 11.84DD366 pKa = 3.4PSDD369 pKa = 3.9PGDD372 pKa = 3.53TRR374 pKa = 11.84EE375 pKa = 4.86PGDD378 pKa = 3.61PCDD381 pKa = 4.01PSEE384 pKa = 4.64EE385 pKa = 4.5GPATEE390 pKa = 4.84SDD392 pKa = 3.75SGSDD396 pKa = 3.04SDD398 pKa = 4.05PNTRR402 pKa = 11.84RR403 pKa = 11.84EE404 pKa = 4.28SPPGPWDD411 pKa = 3.49TRR413 pKa = 11.84PTGPQPPRR421 pKa = 11.84TPEE424 pKa = 3.48EE425 pKa = 3.75AAAAAAATVTPPGAKK440 pKa = 9.03EE441 pKa = 3.24KK442 pKa = 10.42AYY444 pKa = 10.8RR445 pKa = 11.84EE446 pKa = 4.12GRR448 pKa = 11.84GLRR451 pKa = 11.84RR452 pKa = 11.84QRR454 pKa = 11.84PPTPPEE460 pKa = 3.98CYY462 pKa = 9.4RR463 pKa = 11.84RR464 pKa = 11.84VPFTRR469 pKa = 11.84QQSTKK474 pKa = 10.41SRR476 pKa = 11.84NTTLVSPKK484 pKa = 8.64QTPVKK489 pKa = 10.11QGPSSPLAASTPAAATRR506 pKa = 11.84SRR508 pKa = 11.84TTRR511 pKa = 11.84RR512 pKa = 11.84NRR514 pKa = 11.84SRR516 pKa = 11.84SPTNPVASQPVGIAGRR532 pKa = 11.84TRR534 pKa = 11.84RR535 pKa = 11.84SRR537 pKa = 11.84YY538 pKa = 8.21PVEE541 pKa = 4.27EE542 pKa = 4.34EE543 pKa = 4.34SCGGDD548 pKa = 3.13PGGTLQSLRR557 pKa = 11.84DD558 pKa = 3.58PDD560 pKa = 3.55NQRR563 pKa = 11.84EE564 pKa = 4.05QRR566 pKa = 11.84PGQSLPVLGSPAVVLLKK583 pKa = 11.04GGTNQLKK590 pKa = 8.58CCRR593 pKa = 11.84YY594 pKa = 9.45RR595 pKa = 11.84FRR597 pKa = 11.84TRR599 pKa = 11.84HH600 pKa = 3.5QHH602 pKa = 6.71LFLHH606 pKa = 7.08CSSTWYY612 pKa = 9.56WSDD615 pKa = 2.8GNRR618 pKa = 11.84IGRR621 pKa = 11.84SRR623 pKa = 11.84MFLTFRR629 pKa = 11.84DD630 pKa = 3.97PEE632 pKa = 4.05QRR634 pKa = 11.84DD635 pKa = 3.46LFLQTVSIPRR645 pKa = 11.84DD646 pKa = 3.19VDD648 pKa = 3.31IVTGTLFSLL657 pKa = 4.44
Molecular weight: 73.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2534 |
90 |
657 |
422.3 |
47.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.959 ± 0.3 | 2.407 ± 0.631 |
6.117 ± 0.403 | 5.722 ± 0.356 |
3.867 ± 0.511 | 7.103 ± 0.606 |
1.697 ± 0.127 | 4.301 ± 0.545 |
4.538 ± 0.891 | 8.09 ± 0.757 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.618 ± 0.435 | 3.473 ± 0.897 |
7.419 ± 1.133 | 4.657 ± 0.613 |
7.301 ± 1.054 | 8.287 ± 0.889 |
7.064 ± 0.765 | 5.998 ± 0.425 |
1.342 ± 0.337 | 3.039 ± 0.482 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
