
Sanxia tombus-like virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.53
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
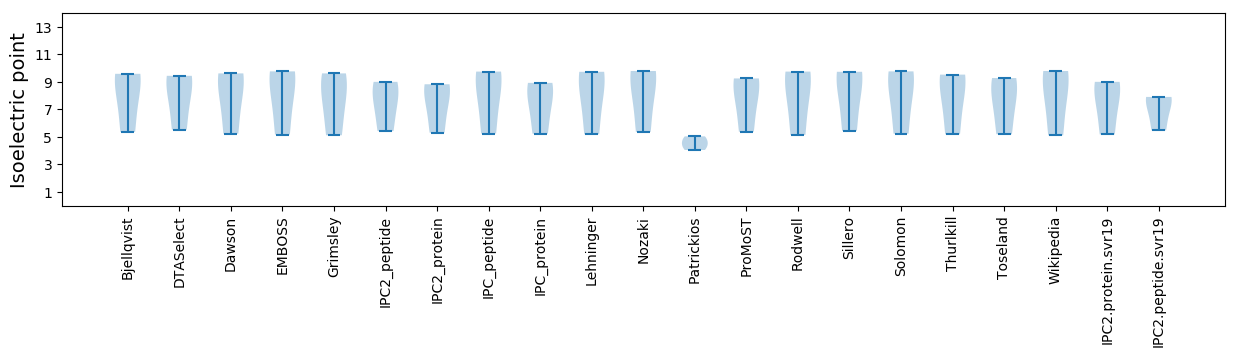
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KGQ8|A0A1L3KGQ8_9VIRU Capsid protein OS=Sanxia tombus-like virus 2 OX=1923386 PE=3 SV=1
MM1 pKa = 7.89AKK3 pKa = 9.66TNQNKK8 pKa = 9.18NKK10 pKa = 9.6PMKK13 pKa = 10.48QNGQVARR20 pKa = 11.84QSASKK25 pKa = 10.42GFTSMGYY32 pKa = 10.53SNNNQDD38 pKa = 3.46PRR40 pKa = 11.84FSNSSGGTVIKK51 pKa = 10.42HH52 pKa = 6.01RR53 pKa = 11.84EE54 pKa = 4.05LIEE57 pKa = 4.14LVSSQTQFNVAQKK70 pKa = 10.29RR71 pKa = 11.84INPADD76 pKa = 3.9RR77 pKa = 11.84ATFPWLSAVAAKK89 pKa = 10.05YY90 pKa = 9.34EE91 pKa = 4.07KK92 pKa = 10.78YY93 pKa = 10.12RR94 pKa = 11.84FRR96 pKa = 11.84KK97 pKa = 9.77LKK99 pKa = 10.28FQMVPHH105 pKa = 7.1APTTASGTLGMYY117 pKa = 9.87IDD119 pKa = 5.16YY120 pKa = 10.83DD121 pKa = 3.92PSDD124 pKa = 3.83VPAPNASAFFSSGNAVTSQIWLEE147 pKa = 4.02TSVQAKK153 pKa = 7.37PQQMQLFNQDD163 pKa = 3.99SIQMADD169 pKa = 5.19LEE171 pKa = 4.89IKK173 pKa = 9.23WFDD176 pKa = 3.31YY177 pKa = 11.21ARR179 pKa = 11.84IFFYY183 pKa = 9.97MQSPGSNGAHH193 pKa = 5.3TAWLFVDD200 pKa = 4.11YY201 pKa = 10.08EE202 pKa = 4.39IEE204 pKa = 4.63LFTPTANVEE213 pKa = 4.28GSKK216 pKa = 8.91FTPDD220 pKa = 3.37FTGFAYY226 pKa = 7.53WTTAQNTAAFTNAPTILRR244 pKa = 11.84GLEE247 pKa = 3.87YY248 pKa = 10.65VQEE251 pKa = 4.5DD252 pKa = 4.61GEE254 pKa = 4.5AKK256 pKa = 9.65LTVPYY261 pKa = 9.98SGYY264 pKa = 10.88YY265 pKa = 10.31RR266 pKa = 11.84ITTLTANTNSGASVTEE282 pKa = 4.02IFEE285 pKa = 4.63SPTCRR290 pKa = 11.84IINGLDD296 pKa = 3.5GEE298 pKa = 4.93GNPTTRR304 pKa = 11.84DD305 pKa = 3.07QTVIVQLQQGEE316 pKa = 4.97TYY318 pKa = 10.71VPLRR322 pKa = 11.84MTTGILDD329 pKa = 4.25LFADD333 pKa = 4.62PDD335 pKa = 4.05NDD337 pKa = 3.84PSPLGVEE344 pKa = 4.4VVLLQASS351 pKa = 3.68
MM1 pKa = 7.89AKK3 pKa = 9.66TNQNKK8 pKa = 9.18NKK10 pKa = 9.6PMKK13 pKa = 10.48QNGQVARR20 pKa = 11.84QSASKK25 pKa = 10.42GFTSMGYY32 pKa = 10.53SNNNQDD38 pKa = 3.46PRR40 pKa = 11.84FSNSSGGTVIKK51 pKa = 10.42HH52 pKa = 6.01RR53 pKa = 11.84EE54 pKa = 4.05LIEE57 pKa = 4.14LVSSQTQFNVAQKK70 pKa = 10.29RR71 pKa = 11.84INPADD76 pKa = 3.9RR77 pKa = 11.84ATFPWLSAVAAKK89 pKa = 10.05YY90 pKa = 9.34EE91 pKa = 4.07KK92 pKa = 10.78YY93 pKa = 10.12RR94 pKa = 11.84FRR96 pKa = 11.84KK97 pKa = 9.77LKK99 pKa = 10.28FQMVPHH105 pKa = 7.1APTTASGTLGMYY117 pKa = 9.87IDD119 pKa = 5.16YY120 pKa = 10.83DD121 pKa = 3.92PSDD124 pKa = 3.83VPAPNASAFFSSGNAVTSQIWLEE147 pKa = 4.02TSVQAKK153 pKa = 7.37PQQMQLFNQDD163 pKa = 3.99SIQMADD169 pKa = 5.19LEE171 pKa = 4.89IKK173 pKa = 9.23WFDD176 pKa = 3.31YY177 pKa = 11.21ARR179 pKa = 11.84IFFYY183 pKa = 9.97MQSPGSNGAHH193 pKa = 5.3TAWLFVDD200 pKa = 4.11YY201 pKa = 10.08EE202 pKa = 4.39IEE204 pKa = 4.63LFTPTANVEE213 pKa = 4.28GSKK216 pKa = 8.91FTPDD220 pKa = 3.37FTGFAYY226 pKa = 7.53WTTAQNTAAFTNAPTILRR244 pKa = 11.84GLEE247 pKa = 3.87YY248 pKa = 10.65VQEE251 pKa = 4.5DD252 pKa = 4.61GEE254 pKa = 4.5AKK256 pKa = 9.65LTVPYY261 pKa = 9.98SGYY264 pKa = 10.88YY265 pKa = 10.31RR266 pKa = 11.84ITTLTANTNSGASVTEE282 pKa = 4.02IFEE285 pKa = 4.63SPTCRR290 pKa = 11.84IINGLDD296 pKa = 3.5GEE298 pKa = 4.93GNPTTRR304 pKa = 11.84DD305 pKa = 3.07QTVIVQLQQGEE316 pKa = 4.97TYY318 pKa = 10.71VPLRR322 pKa = 11.84MTTGILDD329 pKa = 4.25LFADD333 pKa = 4.62PDD335 pKa = 4.05NDD337 pKa = 3.84PSPLGVEE344 pKa = 4.4VVLLQASS351 pKa = 3.68
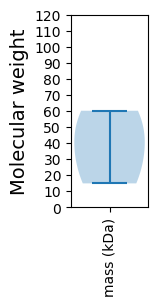
Molecular weight: 38.9 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGQ8|A0A1L3KGQ8_9VIRU Capsid protein OS=Sanxia tombus-like virus 2 OX=1923386 PE=3 SV=1
MM1 pKa = 7.26SRR3 pKa = 11.84VDD5 pKa = 3.41PGIPGGTPLQHH16 pKa = 6.48KK17 pKa = 9.57SSRR20 pKa = 11.84NLMLIIIDD28 pKa = 3.32GHH30 pKa = 7.58IPMVSLMGQSSVILLGAQSRR50 pKa = 11.84SFAFLFVGLTCSTLWKK66 pKa = 9.52KK67 pKa = 10.8VLMRR71 pKa = 11.84YY72 pKa = 9.48FSLMRR77 pKa = 11.84NMSRR81 pKa = 11.84CCSSASAGSKK91 pKa = 8.57QRR93 pKa = 11.84EE94 pKa = 4.27GNLLRR99 pKa = 11.84TCYY102 pKa = 10.18RR103 pKa = 11.84QIFWDD108 pKa = 3.93CLSTGRR114 pKa = 11.84DD115 pKa = 3.54QPSGLPYY122 pKa = 10.77VLDD125 pKa = 3.3WLAWLDD131 pKa = 3.64AGG133 pKa = 5.22
MM1 pKa = 7.26SRR3 pKa = 11.84VDD5 pKa = 3.41PGIPGGTPLQHH16 pKa = 6.48KK17 pKa = 9.57SSRR20 pKa = 11.84NLMLIIIDD28 pKa = 3.32GHH30 pKa = 7.58IPMVSLMGQSSVILLGAQSRR50 pKa = 11.84SFAFLFVGLTCSTLWKK66 pKa = 9.52KK67 pKa = 10.8VLMRR71 pKa = 11.84YY72 pKa = 9.48FSLMRR77 pKa = 11.84NMSRR81 pKa = 11.84CCSSASAGSKK91 pKa = 8.57QRR93 pKa = 11.84EE94 pKa = 4.27GNLLRR99 pKa = 11.84TCYY102 pKa = 10.18RR103 pKa = 11.84QIFWDD108 pKa = 3.93CLSTGRR114 pKa = 11.84DD115 pKa = 3.54QPSGLPYY122 pKa = 10.77VLDD125 pKa = 3.3WLAWLDD131 pKa = 3.64AGG133 pKa = 5.22
Molecular weight: 14.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1008 |
133 |
524 |
336.0 |
37.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.448 ± 1.234 | 1.984 ± 0.889 |
4.365 ± 0.274 | 4.96 ± 1.101 |
5.258 ± 0.364 | 6.944 ± 0.518 |
1.786 ± 0.534 | 4.464 ± 0.214 |
4.663 ± 0.507 | 8.532 ± 1.423 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.075 ± 0.511 | 4.464 ± 0.958 |
5.06 ± 0.479 | 5.853 ± 0.531 |
6.349 ± 1.381 | 7.937 ± 1.129 |
5.754 ± 1.894 | 6.845 ± 0.964 |
1.786 ± 0.297 | 3.472 ± 0.335 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
