
Pseudogymnoascus sp. VKM F-4520 (FW-2644)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Leotiomycetes; Leotiomycetes incertae sedis; Pseudeurotiaceae; Pseudogymnoascus; unclassified Pseudogymnoascus
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins
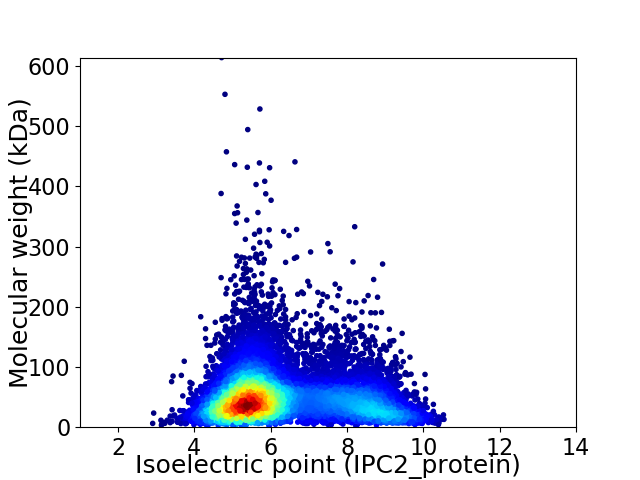
Virtual 2D-PAGE plot for 11550 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A094HH73|A0A094HH73_9PEZI Ubiquitin carboxyl-terminal hydrolase OS=Pseudogymnoascus sp. VKM F-4520 (FW-2644) OX=1420915 GN=V502_06173 PE=3 SV=1
MM1 pKa = 7.05VAIKK5 pKa = 10.14PVFALSLLAGLITPALSAAVKK26 pKa = 10.17INAFGDD32 pKa = 4.24SITGSPGCWRR42 pKa = 11.84ALLYY46 pKa = 10.54QKK48 pKa = 10.54LVEE51 pKa = 5.28AGTTDD56 pKa = 2.88IDD58 pKa = 4.5FVGTLPGQGCGIEE71 pKa = 3.98YY72 pKa = 10.6DD73 pKa = 4.23GEE75 pKa = 4.34NDD77 pKa = 3.27GHH79 pKa = 7.39GGFLATGIVADD90 pKa = 3.97NQLPGWLAISQPDD103 pKa = 3.44IVMMQLATNDD113 pKa = 2.73IWSNIATATILDD125 pKa = 4.09AFSALVDD132 pKa = 3.29QMRR135 pKa = 11.84DD136 pKa = 3.28SKK138 pKa = 10.74STMHH142 pKa = 6.54IVVAQITPMDD152 pKa = 4.24PTDD155 pKa = 3.45GCATCEE161 pKa = 3.88AGIIALNDD169 pKa = 5.02AIPAWVAEE177 pKa = 4.17KK178 pKa = 9.96STIEE182 pKa = 4.04SPITVVDD189 pKa = 4.99CYY191 pKa = 10.57TGYY194 pKa = 8.59DD195 pKa = 3.85TATDD199 pKa = 3.96TYY201 pKa = 11.53DD202 pKa = 3.5GVHH205 pKa = 6.79PNDD208 pKa = 3.71SGNVKK213 pKa = 10.25LADD216 pKa = 3.21AWFEE220 pKa = 3.91PLQAAILAASSGTNSTIAA238 pKa = 3.51
MM1 pKa = 7.05VAIKK5 pKa = 10.14PVFALSLLAGLITPALSAAVKK26 pKa = 10.17INAFGDD32 pKa = 4.24SITGSPGCWRR42 pKa = 11.84ALLYY46 pKa = 10.54QKK48 pKa = 10.54LVEE51 pKa = 5.28AGTTDD56 pKa = 2.88IDD58 pKa = 4.5FVGTLPGQGCGIEE71 pKa = 3.98YY72 pKa = 10.6DD73 pKa = 4.23GEE75 pKa = 4.34NDD77 pKa = 3.27GHH79 pKa = 7.39GGFLATGIVADD90 pKa = 3.97NQLPGWLAISQPDD103 pKa = 3.44IVMMQLATNDD113 pKa = 2.73IWSNIATATILDD125 pKa = 4.09AFSALVDD132 pKa = 3.29QMRR135 pKa = 11.84DD136 pKa = 3.28SKK138 pKa = 10.74STMHH142 pKa = 6.54IVVAQITPMDD152 pKa = 4.24PTDD155 pKa = 3.45GCATCEE161 pKa = 3.88AGIIALNDD169 pKa = 5.02AIPAWVAEE177 pKa = 4.17KK178 pKa = 9.96STIEE182 pKa = 4.04SPITVVDD189 pKa = 4.99CYY191 pKa = 10.57TGYY194 pKa = 8.59DD195 pKa = 3.85TATDD199 pKa = 3.96TYY201 pKa = 11.53DD202 pKa = 3.5GVHH205 pKa = 6.79PNDD208 pKa = 3.71SGNVKK213 pKa = 10.25LADD216 pKa = 3.21AWFEE220 pKa = 3.91PLQAAILAASSGTNSTIAA238 pKa = 3.51
Molecular weight: 24.78 kDa
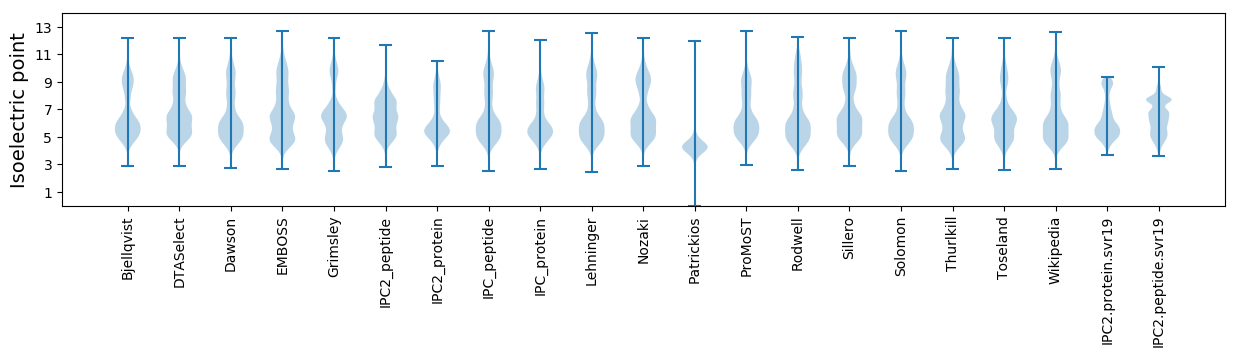
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A094HZ68|A0A094HZ68_9PEZI CUE domain-containing protein OS=Pseudogymnoascus sp. VKM F-4520 (FW-2644) OX=1420915 GN=V502_03319 PE=4 SV=1
AA1 pKa = 7.98DD2 pKa = 3.92SEE4 pKa = 4.62NLVLAADD11 pKa = 4.0PADD14 pKa = 3.86RR15 pKa = 11.84ATSRR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84KK22 pKa = 10.04LILLACWVPSSPDD35 pKa = 3.32DD36 pKa = 4.04GEE38 pKa = 4.41AAGSRR43 pKa = 11.84SRR45 pKa = 11.84AARR48 pKa = 11.84MVSWGVVMGDD58 pKa = 3.29STADD62 pKa = 3.32TDD64 pKa = 3.66AGSAKK69 pKa = 10.48GPTEE73 pKa = 3.38KK74 pKa = 10.06RR75 pKa = 11.84RR76 pKa = 11.84EE77 pKa = 3.82GRR79 pKa = 11.84RR80 pKa = 11.84RR81 pKa = 11.84GMQGMSKK88 pKa = 10.43GSWARR93 pKa = 11.84RR94 pKa = 11.84TQVIGADD101 pKa = 3.49TSAEE105 pKa = 3.97EE106 pKa = 4.17RR107 pKa = 11.84GVEE110 pKa = 4.17LMQRR114 pKa = 11.84SLGRR118 pKa = 11.84HH119 pKa = 5.12EE120 pKa = 5.72LSTALRR126 pKa = 11.84NPASEE131 pKa = 4.09RR132 pKa = 11.84RR133 pKa = 11.84LPASATRR140 pKa = 11.84VQYY143 pKa = 11.48DD144 pKa = 3.56DD145 pKa = 4.61ASSHH149 pKa = 5.99GSADD153 pKa = 3.48LLRR156 pKa = 11.84RR157 pKa = 11.84VADD160 pKa = 3.6VAGGSAGGVAGGRR173 pKa = 11.84RR174 pKa = 11.84AANGRR179 pKa = 11.84LWGGGTGGGGSYY191 pKa = 10.6CGG193 pKa = 3.64
AA1 pKa = 7.98DD2 pKa = 3.92SEE4 pKa = 4.62NLVLAADD11 pKa = 4.0PADD14 pKa = 3.86RR15 pKa = 11.84ATSRR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84KK22 pKa = 10.04LILLACWVPSSPDD35 pKa = 3.32DD36 pKa = 4.04GEE38 pKa = 4.41AAGSRR43 pKa = 11.84SRR45 pKa = 11.84AARR48 pKa = 11.84MVSWGVVMGDD58 pKa = 3.29STADD62 pKa = 3.32TDD64 pKa = 3.66AGSAKK69 pKa = 10.48GPTEE73 pKa = 3.38KK74 pKa = 10.06RR75 pKa = 11.84RR76 pKa = 11.84EE77 pKa = 3.82GRR79 pKa = 11.84RR80 pKa = 11.84RR81 pKa = 11.84GMQGMSKK88 pKa = 10.43GSWARR93 pKa = 11.84RR94 pKa = 11.84TQVIGADD101 pKa = 3.49TSAEE105 pKa = 3.97EE106 pKa = 4.17RR107 pKa = 11.84GVEE110 pKa = 4.17LMQRR114 pKa = 11.84SLGRR118 pKa = 11.84HH119 pKa = 5.12EE120 pKa = 5.72LSTALRR126 pKa = 11.84NPASEE131 pKa = 4.09RR132 pKa = 11.84RR133 pKa = 11.84LPASATRR140 pKa = 11.84VQYY143 pKa = 11.48DD144 pKa = 3.56DD145 pKa = 4.61ASSHH149 pKa = 5.99GSADD153 pKa = 3.48LLRR156 pKa = 11.84RR157 pKa = 11.84VADD160 pKa = 3.6VAGGSAGGVAGGRR173 pKa = 11.84RR174 pKa = 11.84AANGRR179 pKa = 11.84LWGGGTGGGGSYY191 pKa = 10.6CGG193 pKa = 3.64
Molecular weight: 20.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5669913 |
10 |
5591 |
490.9 |
54.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.923 ± 0.023 | 1.219 ± 0.007 |
5.642 ± 0.015 | 6.258 ± 0.023 |
3.657 ± 0.013 | 7.255 ± 0.023 |
2.272 ± 0.009 | 5.138 ± 0.015 |
4.928 ± 0.018 | 8.807 ± 0.022 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.145 ± 0.008 | 3.745 ± 0.011 |
5.834 ± 0.022 | 3.866 ± 0.017 |
5.792 ± 0.019 | 8.036 ± 0.026 |
6.053 ± 0.017 | 6.196 ± 0.015 |
1.452 ± 0.008 | 2.781 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
