
Acidiphilium rubrum
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Acetobacteraceae; Acidiphilium
Average proteome isoelectric point is 6.99
Get precalculated fractions of proteins
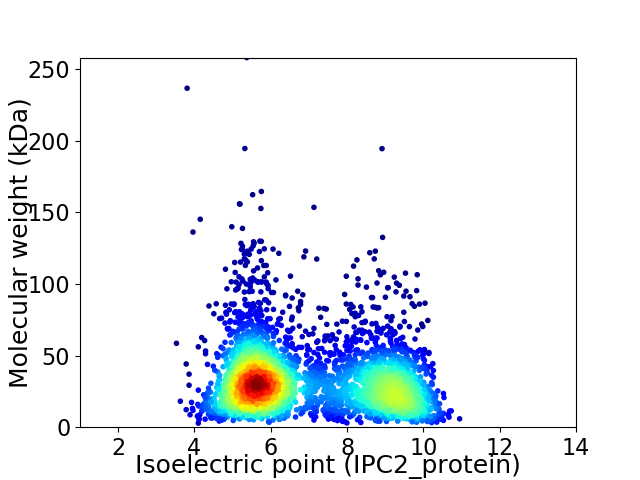
Virtual 2D-PAGE plot for 3677 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1N6Z510|A0A1N6Z510_ACIRU tRNA-dihydrouridine synthase OS=Acidiphilium rubrum OX=526 GN=SAMN05421828_1202 PE=3 SV=1
MM1 pKa = 7.59SIFGALDD8 pKa = 3.41TSVSGMQAQSSAFTNISDD26 pKa = 5.06NIANSQTTGYY36 pKa = 10.38KK37 pKa = 10.56GVDD40 pKa = 3.34TNFINYY46 pKa = 8.68LVQSSPSSNGADD58 pKa = 3.1SVIARR63 pKa = 11.84PSYY66 pKa = 9.57TNTVQGTVTQSTNPLALAISGQGFFNVSEE95 pKa = 4.26PAASATAGGTTQSFQSQQYY114 pKa = 6.0YY115 pKa = 8.8TRR117 pKa = 11.84AGDD120 pKa = 3.5FTLNKK125 pKa = 9.68QGYY128 pKa = 8.56LVNSAGAYY136 pKa = 9.54LNAWPIDD143 pKa = 3.66TATGVVNTSSLAPIQISQATTPPVATSSVSLSASLPTTPATSPVNTQVDD192 pKa = 3.86IYY194 pKa = 11.36DD195 pKa = 3.66SLGNLQQLNLSWTQTGTNQWNLAVYY220 pKa = 10.45APGDD224 pKa = 3.9PGGSTAKK231 pKa = 10.74APIANASMTFGSDD244 pKa = 3.02GTLSALSTTTTNAAASANTAGQPATLTIPANFGNGTQNIALDD286 pKa = 3.98FGRR289 pKa = 11.84FGGTAGLTQYY299 pKa = 11.54AGTSLNLQGASQNGTPPGNFSNLSIDD325 pKa = 3.53TQGNITVNYY334 pKa = 10.37NNGFSQAVAQIPIATFDD351 pKa = 5.06APDD354 pKa = 3.73ALQNQSGQIYY364 pKa = 7.79TASQGSGSATINPVGSNGNALVTGSTEE391 pKa = 3.89SSNVDD396 pKa = 2.62IATQFTNLITAQQAYY411 pKa = 5.86TANSKK416 pKa = 10.9VITTAQQLLTTVVNMVQQ433 pKa = 2.91
MM1 pKa = 7.59SIFGALDD8 pKa = 3.41TSVSGMQAQSSAFTNISDD26 pKa = 5.06NIANSQTTGYY36 pKa = 10.38KK37 pKa = 10.56GVDD40 pKa = 3.34TNFINYY46 pKa = 8.68LVQSSPSSNGADD58 pKa = 3.1SVIARR63 pKa = 11.84PSYY66 pKa = 9.57TNTVQGTVTQSTNPLALAISGQGFFNVSEE95 pKa = 4.26PAASATAGGTTQSFQSQQYY114 pKa = 6.0YY115 pKa = 8.8TRR117 pKa = 11.84AGDD120 pKa = 3.5FTLNKK125 pKa = 9.68QGYY128 pKa = 8.56LVNSAGAYY136 pKa = 9.54LNAWPIDD143 pKa = 3.66TATGVVNTSSLAPIQISQATTPPVATSSVSLSASLPTTPATSPVNTQVDD192 pKa = 3.86IYY194 pKa = 11.36DD195 pKa = 3.66SLGNLQQLNLSWTQTGTNQWNLAVYY220 pKa = 10.45APGDD224 pKa = 3.9PGGSTAKK231 pKa = 10.74APIANASMTFGSDD244 pKa = 3.02GTLSALSTTTTNAAASANTAGQPATLTIPANFGNGTQNIALDD286 pKa = 3.98FGRR289 pKa = 11.84FGGTAGLTQYY299 pKa = 11.54AGTSLNLQGASQNGTPPGNFSNLSIDD325 pKa = 3.53TQGNITVNYY334 pKa = 10.37NNGFSQAVAQIPIATFDD351 pKa = 5.06APDD354 pKa = 3.73ALQNQSGQIYY364 pKa = 7.79TASQGSGSATINPVGSNGNALVTGSTEE391 pKa = 3.89SSNVDD396 pKa = 2.62IATQFTNLITAQQAYY411 pKa = 5.86TANSKK416 pKa = 10.9VITTAQQLLTTVVNMVQQ433 pKa = 2.91
Molecular weight: 44.13 kDa
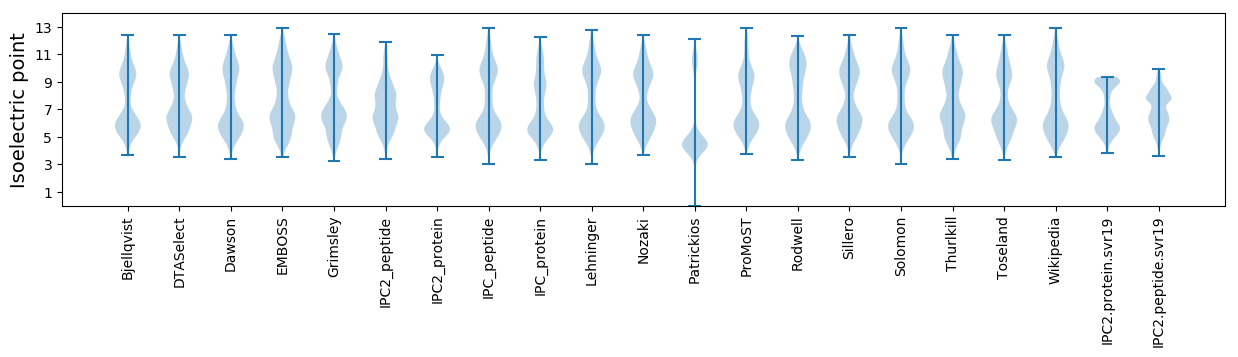
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1N7BBQ8|A0A1N7BBQ8_ACIRU Peptide/nickel transport system ATP-binding protein OS=Acidiphilium rubrum OX=526 GN=SAMN05421828_13815 PE=4 SV=1
FF1 pKa = 6.72YY2 pKa = 10.34RR3 pKa = 11.84ISAKK7 pKa = 10.27RR8 pKa = 11.84GSLPEE13 pKa = 4.06NLTGIVVHH21 pKa = 6.31DD22 pKa = 3.67HH23 pKa = 6.04WKK25 pKa = 10.09PYY27 pKa = 8.06YY28 pKa = 8.73TLAGVLHH35 pKa = 6.76ALCNAHH41 pKa = 7.15HH42 pKa = 6.41LRR44 pKa = 11.84EE45 pKa = 4.15LQALIEE51 pKa = 4.4IEE53 pKa = 4.1HH54 pKa = 6.7EE55 pKa = 3.73DD56 pKa = 2.98WARR59 pKa = 11.84RR60 pKa = 11.84MQTLLRR66 pKa = 11.84RR67 pKa = 11.84ACHH70 pKa = 5.87AANLSRR76 pKa = 11.84MQGKK80 pKa = 8.44PLPPRR85 pKa = 11.84LIALIEE91 pKa = 3.96RR92 pKa = 11.84CYY94 pKa = 11.02DD95 pKa = 4.07AIVTDD100 pKa = 4.18GLLFHH105 pKa = 7.15TDD107 pKa = 3.48QSPLTQADD115 pKa = 3.38VQARR119 pKa = 11.84RR120 pKa = 11.84RR121 pKa = 11.84GRR123 pKa = 11.84VPRR126 pKa = 11.84RR127 pKa = 11.84VGHH130 pKa = 6.25NLLLRR135 pKa = 11.84LSTRR139 pKa = 11.84KK140 pKa = 9.49PDD142 pKa = 3.5VLRR145 pKa = 11.84FLSNPAVPFTNNLAEE160 pKa = 4.46RR161 pKa = 11.84DD162 pKa = 3.6GRR164 pKa = 11.84MMKK167 pKa = 10.34LRR169 pKa = 11.84QKK171 pKa = 10.45ISGGFRR177 pKa = 11.84AVTGAEE183 pKa = 3.93DD184 pKa = 3.6FAVIRR189 pKa = 11.84SFFSTAKK196 pKa = 9.12KK197 pKa = 9.16QQWDD201 pKa = 3.5VLNAINAHH209 pKa = 6.0PNVLIAQLKK218 pKa = 8.76LAA220 pKa = 4.7
FF1 pKa = 6.72YY2 pKa = 10.34RR3 pKa = 11.84ISAKK7 pKa = 10.27RR8 pKa = 11.84GSLPEE13 pKa = 4.06NLTGIVVHH21 pKa = 6.31DD22 pKa = 3.67HH23 pKa = 6.04WKK25 pKa = 10.09PYY27 pKa = 8.06YY28 pKa = 8.73TLAGVLHH35 pKa = 6.76ALCNAHH41 pKa = 7.15HH42 pKa = 6.41LRR44 pKa = 11.84EE45 pKa = 4.15LQALIEE51 pKa = 4.4IEE53 pKa = 4.1HH54 pKa = 6.7EE55 pKa = 3.73DD56 pKa = 2.98WARR59 pKa = 11.84RR60 pKa = 11.84MQTLLRR66 pKa = 11.84RR67 pKa = 11.84ACHH70 pKa = 5.87AANLSRR76 pKa = 11.84MQGKK80 pKa = 8.44PLPPRR85 pKa = 11.84LIALIEE91 pKa = 3.96RR92 pKa = 11.84CYY94 pKa = 11.02DD95 pKa = 4.07AIVTDD100 pKa = 4.18GLLFHH105 pKa = 7.15TDD107 pKa = 3.48QSPLTQADD115 pKa = 3.38VQARR119 pKa = 11.84RR120 pKa = 11.84RR121 pKa = 11.84GRR123 pKa = 11.84VPRR126 pKa = 11.84RR127 pKa = 11.84VGHH130 pKa = 6.25NLLLRR135 pKa = 11.84LSTRR139 pKa = 11.84KK140 pKa = 9.49PDD142 pKa = 3.5VLRR145 pKa = 11.84FLSNPAVPFTNNLAEE160 pKa = 4.46RR161 pKa = 11.84DD162 pKa = 3.6GRR164 pKa = 11.84MMKK167 pKa = 10.34LRR169 pKa = 11.84QKK171 pKa = 10.45ISGGFRR177 pKa = 11.84AVTGAEE183 pKa = 3.93DD184 pKa = 3.6FAVIRR189 pKa = 11.84SFFSTAKK196 pKa = 9.12KK197 pKa = 9.16QQWDD201 pKa = 3.5VLNAINAHH209 pKa = 6.0PNVLIAQLKK218 pKa = 8.76LAA220 pKa = 4.7
Molecular weight: 25.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1184703 |
26 |
2478 |
322.2 |
34.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.203 ± 0.066 | 0.842 ± 0.011 |
5.429 ± 0.029 | 4.389 ± 0.042 |
3.588 ± 0.025 | 8.715 ± 0.044 |
2.349 ± 0.021 | 5.778 ± 0.026 |
2.529 ± 0.031 | 10.355 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.443 ± 0.017 | 2.513 ± 0.025 |
5.712 ± 0.04 | 3.028 ± 0.025 |
7.128 ± 0.048 | 4.933 ± 0.029 |
5.753 ± 0.033 | 6.943 ± 0.031 |
1.278 ± 0.018 | 2.09 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
