
Microbacterium phage Araxxi
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Podoviridae; Burrovirus; Microbacterium virus Araxxi
Average proteome isoelectric point is 5.77
Get precalculated fractions of proteins
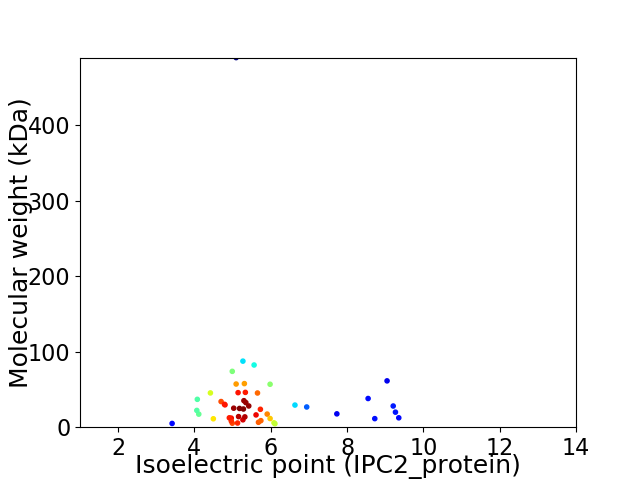
Virtual 2D-PAGE plot for 50 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
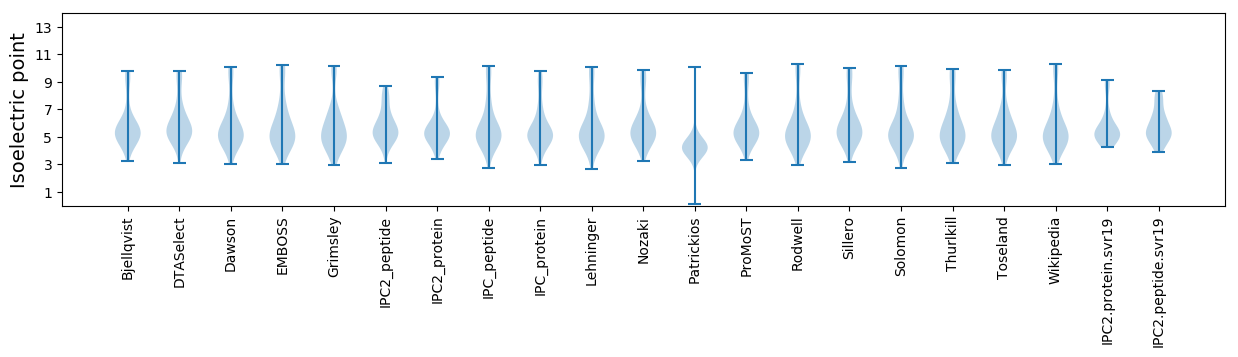
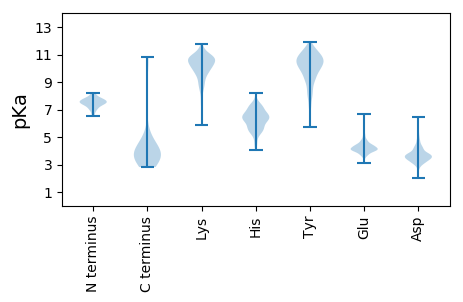
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A516KT73|A0A516KT73_9CAUD Uncharacterized protein OS=Microbacterium phage Araxxi OX=2590948 GN=41 PE=4 SV=1
MM1 pKa = 7.85SDD3 pKa = 3.06INEE6 pKa = 4.28NTDD9 pKa = 3.52GLLTEE14 pKa = 4.69QDD16 pKa = 3.68QADD19 pKa = 4.34LAQLAAEE26 pKa = 4.17PLEE29 pKa = 4.15ITVLEE34 pKa = 4.24TWEE37 pKa = 4.4SVLSNIEE44 pKa = 3.57QMEE47 pKa = 4.11NEE49 pKa = 4.45KK50 pKa = 10.35IEE52 pKa = 3.83PSYY55 pKa = 11.28AVNIVGKK62 pKa = 9.08WPKK65 pKa = 10.2LDD67 pKa = 3.96YY68 pKa = 11.36GDD70 pKa = 4.15VPHH73 pKa = 7.27FYY75 pKa = 11.23KK76 pKa = 10.98LFNEE80 pKa = 3.89YY81 pKa = 10.25LKK83 pKa = 10.41RR84 pKa = 11.84YY85 pKa = 9.0RR86 pKa = 11.84AVLEE90 pKa = 4.1EE91 pKa = 3.86QLRR94 pKa = 11.84LHH96 pKa = 7.31PDD98 pKa = 2.96AKK100 pKa = 11.13DD101 pKa = 3.28NTGEE105 pKa = 4.12FGTEE109 pKa = 3.62EE110 pKa = 3.88SDD112 pKa = 4.43AIANRR117 pKa = 11.84DD118 pKa = 3.18IYY120 pKa = 11.03VEE122 pKa = 5.8LMFGWNVVTAEE133 pKa = 5.4LEE135 pKa = 4.46HH136 pKa = 6.67EE137 pKa = 4.06WDD139 pKa = 4.14ALDD142 pKa = 3.33VDD144 pKa = 4.23AAEE147 pKa = 5.4KK148 pKa = 9.5IAAMGEE154 pKa = 3.95AQLFVTGPQGMLQALTAPNVGFQWTNEE181 pKa = 3.84EE182 pKa = 4.11QIALTEE188 pKa = 4.11RR189 pKa = 11.84VVAAAEE195 pKa = 4.04EE196 pKa = 4.28LL197 pKa = 3.84
MM1 pKa = 7.85SDD3 pKa = 3.06INEE6 pKa = 4.28NTDD9 pKa = 3.52GLLTEE14 pKa = 4.69QDD16 pKa = 3.68QADD19 pKa = 4.34LAQLAAEE26 pKa = 4.17PLEE29 pKa = 4.15ITVLEE34 pKa = 4.24TWEE37 pKa = 4.4SVLSNIEE44 pKa = 3.57QMEE47 pKa = 4.11NEE49 pKa = 4.45KK50 pKa = 10.35IEE52 pKa = 3.83PSYY55 pKa = 11.28AVNIVGKK62 pKa = 9.08WPKK65 pKa = 10.2LDD67 pKa = 3.96YY68 pKa = 11.36GDD70 pKa = 4.15VPHH73 pKa = 7.27FYY75 pKa = 11.23KK76 pKa = 10.98LFNEE80 pKa = 3.89YY81 pKa = 10.25LKK83 pKa = 10.41RR84 pKa = 11.84YY85 pKa = 9.0RR86 pKa = 11.84AVLEE90 pKa = 4.1EE91 pKa = 3.86QLRR94 pKa = 11.84LHH96 pKa = 7.31PDD98 pKa = 2.96AKK100 pKa = 11.13DD101 pKa = 3.28NTGEE105 pKa = 4.12FGTEE109 pKa = 3.62EE110 pKa = 3.88SDD112 pKa = 4.43AIANRR117 pKa = 11.84DD118 pKa = 3.18IYY120 pKa = 11.03VEE122 pKa = 5.8LMFGWNVVTAEE133 pKa = 5.4LEE135 pKa = 4.46HH136 pKa = 6.67EE137 pKa = 4.06WDD139 pKa = 4.14ALDD142 pKa = 3.33VDD144 pKa = 4.23AAEE147 pKa = 5.4KK148 pKa = 9.5IAAMGEE154 pKa = 3.95AQLFVTGPQGMLQALTAPNVGFQWTNEE181 pKa = 3.84EE182 pKa = 4.11QIALTEE188 pKa = 4.11RR189 pKa = 11.84VVAAAEE195 pKa = 4.04EE196 pKa = 4.28LL197 pKa = 3.84
Molecular weight: 22.22 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A516KT34|A0A516KT34_9CAUD Uncharacterized protein OS=Microbacterium phage Araxxi OX=2590948 GN=26 PE=4 SV=1
MM1 pKa = 6.84NQEE4 pKa = 4.4LKK6 pKa = 11.08DD7 pKa = 3.7PFAGSRR13 pKa = 11.84LPSDD17 pKa = 3.78AAAPPQPEE25 pKa = 4.74RR26 pKa = 11.84YY27 pKa = 9.42DD28 pKa = 3.75QLFLPNDD35 pKa = 3.68EE36 pKa = 4.65KK37 pKa = 11.27PRR39 pKa = 11.84MPLTKK44 pKa = 9.86DD45 pKa = 2.8RR46 pKa = 11.84YY47 pKa = 8.12VVRR50 pKa = 11.84DD51 pKa = 3.83NPHH54 pKa = 4.81EE55 pKa = 4.32VAWEE59 pKa = 4.06RR60 pKa = 11.84QVRR63 pKa = 11.84MYY65 pKa = 11.16LRR67 pKa = 11.84GLSLDD72 pKa = 3.71DD73 pKa = 3.54SHH75 pKa = 6.38RR76 pKa = 11.84TSATMIYY83 pKa = 9.85EE84 pKa = 4.04WATGKK89 pKa = 10.28RR90 pKa = 11.84VADD93 pKa = 3.89QIAAGVATSPDD104 pKa = 3.09LRR106 pKa = 11.84KK107 pKa = 9.6INKK110 pKa = 8.92IMRR113 pKa = 11.84FYY115 pKa = 10.75FGKK118 pKa = 9.85PYY120 pKa = 8.65QTWIAGRR127 pKa = 11.84KK128 pKa = 8.74IPNCYY133 pKa = 9.9RR134 pKa = 11.84IPKK137 pKa = 7.65GWRR140 pKa = 11.84VKK142 pKa = 8.85RR143 pKa = 11.84HH144 pKa = 5.56RR145 pKa = 11.84PMTLTLWSEE154 pKa = 4.26YY155 pKa = 10.79TEE157 pKa = 4.22GTLNDD162 pKa = 3.62DD163 pKa = 3.73GARR166 pKa = 11.84RR167 pKa = 11.84NAAQQ171 pKa = 3.38
MM1 pKa = 6.84NQEE4 pKa = 4.4LKK6 pKa = 11.08DD7 pKa = 3.7PFAGSRR13 pKa = 11.84LPSDD17 pKa = 3.78AAAPPQPEE25 pKa = 4.74RR26 pKa = 11.84YY27 pKa = 9.42DD28 pKa = 3.75QLFLPNDD35 pKa = 3.68EE36 pKa = 4.65KK37 pKa = 11.27PRR39 pKa = 11.84MPLTKK44 pKa = 9.86DD45 pKa = 2.8RR46 pKa = 11.84YY47 pKa = 8.12VVRR50 pKa = 11.84DD51 pKa = 3.83NPHH54 pKa = 4.81EE55 pKa = 4.32VAWEE59 pKa = 4.06RR60 pKa = 11.84QVRR63 pKa = 11.84MYY65 pKa = 11.16LRR67 pKa = 11.84GLSLDD72 pKa = 3.71DD73 pKa = 3.54SHH75 pKa = 6.38RR76 pKa = 11.84TSATMIYY83 pKa = 9.85EE84 pKa = 4.04WATGKK89 pKa = 10.28RR90 pKa = 11.84VADD93 pKa = 3.89QIAAGVATSPDD104 pKa = 3.09LRR106 pKa = 11.84KK107 pKa = 9.6INKK110 pKa = 8.92IMRR113 pKa = 11.84FYY115 pKa = 10.75FGKK118 pKa = 9.85PYY120 pKa = 8.65QTWIAGRR127 pKa = 11.84KK128 pKa = 8.74IPNCYY133 pKa = 9.9RR134 pKa = 11.84IPKK137 pKa = 7.65GWRR140 pKa = 11.84VKK142 pKa = 8.85RR143 pKa = 11.84HH144 pKa = 5.56RR145 pKa = 11.84PMTLTLWSEE154 pKa = 4.26YY155 pKa = 10.79TEE157 pKa = 4.22GTLNDD162 pKa = 3.62DD163 pKa = 3.73GARR166 pKa = 11.84RR167 pKa = 11.84NAAQQ171 pKa = 3.38
Molecular weight: 19.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
16885 |
43 |
4494 |
337.7 |
37.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.104 ± 0.534 | 0.444 ± 0.166 |
6.574 ± 0.227 | 5.988 ± 0.343 |
3.311 ± 0.141 | 7.77 ± 0.446 |
1.617 ± 0.223 | 4.625 ± 0.184 |
4.246 ± 0.362 | 8.22 ± 0.299 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.588 ± 0.168 | 4.661 ± 0.279 |
4.904 ± 0.2 | 4.762 ± 0.199 |
5.964 ± 0.321 | 6.189 ± 0.275 |
6.574 ± 0.338 | 6.532 ± 0.241 |
1.836 ± 0.127 | 3.092 ± 0.179 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
