
Pseudomonas lundensis
Taxonomy: cellular organisms; Bacteria;
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
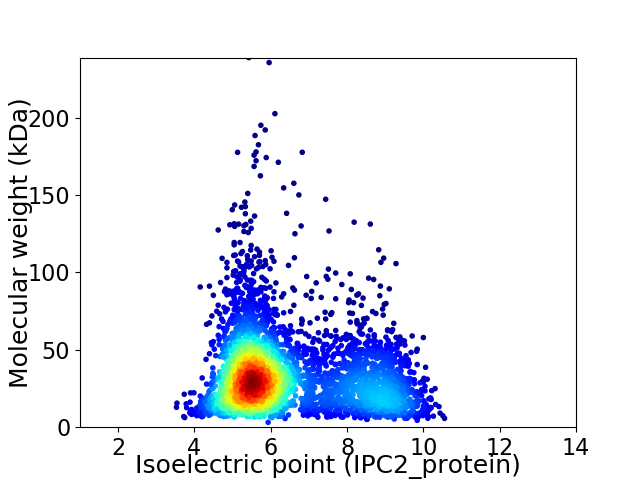
Virtual 2D-PAGE plot for 3977 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
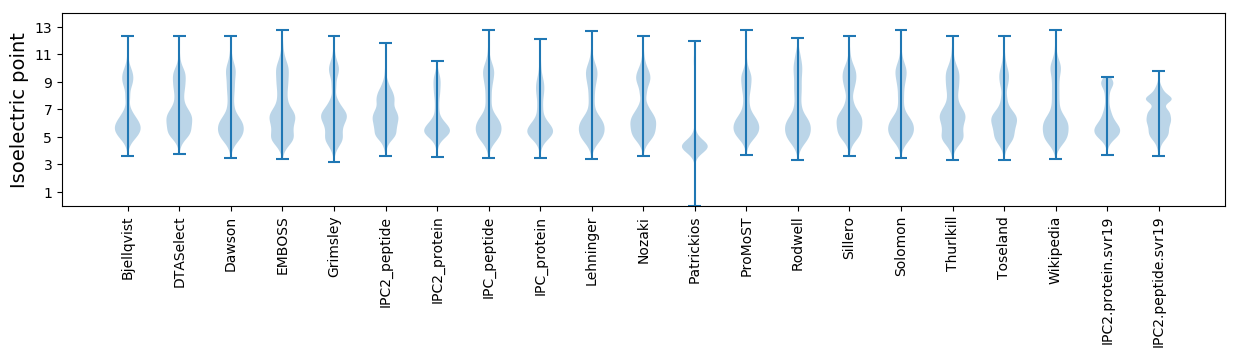
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1D9IJV2|A0A1D9IJV2_9PSED DEAD/DEAH box helicase OS=Pseudomonas lundensis OX=86185 GN=AA042_06585 PE=3 SV=1
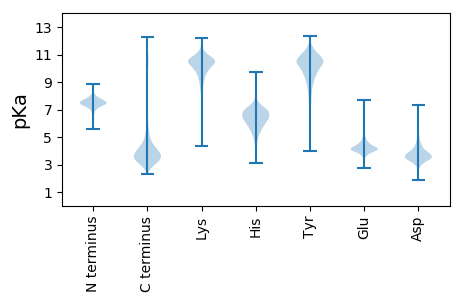
MM1 pKa = 7.52TISITYY7 pKa = 9.73NSAFGAFSVQDD18 pKa = 4.14ALSAWSTGFTTAGHH32 pKa = 5.84GTGNTGGFNTGGRR45 pKa = 11.84AMTGEE50 pKa = 4.26QYY52 pKa = 9.45ATHH55 pKa = 6.9GAQGSEE61 pKa = 4.23YY62 pKa = 10.82AFIAASDD69 pKa = 3.65TDD71 pKa = 3.16KK72 pKa = 11.44GLNYY76 pKa = 9.98VFDD79 pKa = 4.81PKK81 pKa = 10.96VSPGDD86 pKa = 3.71NMNHH90 pKa = 5.55YY91 pKa = 10.47LWGEE95 pKa = 4.13LDD97 pKa = 3.73TVTLGTGLQGGNGTDD112 pKa = 4.1FGLNDD117 pKa = 3.36YY118 pKa = 7.45TASFNGLDD126 pKa = 3.92LSAASDD132 pKa = 3.32AGRR135 pKa = 11.84AGNAVQDD142 pKa = 3.96VIYY145 pKa = 10.67GLMKK149 pKa = 10.6GDD151 pKa = 3.52VSGLEE156 pKa = 3.96GALNNLLSGFGLSTDD171 pKa = 3.4STFDD175 pKa = 3.53DD176 pKa = 4.2LAAAGLAHH184 pKa = 6.92TDD186 pKa = 3.18APLAADD192 pKa = 3.42IDD194 pKa = 4.27LVGVQEE200 pKa = 4.75VAQDD204 pKa = 3.19WALAAA209 pKa = 4.59
MM1 pKa = 7.52TISITYY7 pKa = 9.73NSAFGAFSVQDD18 pKa = 4.14ALSAWSTGFTTAGHH32 pKa = 5.84GTGNTGGFNTGGRR45 pKa = 11.84AMTGEE50 pKa = 4.26QYY52 pKa = 9.45ATHH55 pKa = 6.9GAQGSEE61 pKa = 4.23YY62 pKa = 10.82AFIAASDD69 pKa = 3.65TDD71 pKa = 3.16KK72 pKa = 11.44GLNYY76 pKa = 9.98VFDD79 pKa = 4.81PKK81 pKa = 10.96VSPGDD86 pKa = 3.71NMNHH90 pKa = 5.55YY91 pKa = 10.47LWGEE95 pKa = 4.13LDD97 pKa = 3.73TVTLGTGLQGGNGTDD112 pKa = 4.1FGLNDD117 pKa = 3.36YY118 pKa = 7.45TASFNGLDD126 pKa = 3.92LSAASDD132 pKa = 3.32AGRR135 pKa = 11.84AGNAVQDD142 pKa = 3.96VIYY145 pKa = 10.67GLMKK149 pKa = 10.6GDD151 pKa = 3.52VSGLEE156 pKa = 3.96GALNNLLSGFGLSTDD171 pKa = 3.4STFDD175 pKa = 3.53DD176 pKa = 4.2LAAAGLAHH184 pKa = 6.92TDD186 pKa = 3.18APLAADD192 pKa = 3.42IDD194 pKa = 4.27LVGVQEE200 pKa = 4.75VAQDD204 pKa = 3.19WALAAA209 pKa = 4.59
Molecular weight: 21.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1D9IMR0|A0A1D9IMR0_9PSED Aromatic amino acid lyase OS=Pseudomonas lundensis OX=86185 GN=AA042_12175 PE=4 SV=1
MM1 pKa = 8.01DD2 pKa = 6.19LATQPVSTRR11 pKa = 11.84RR12 pKa = 11.84DD13 pKa = 2.99AWCWRR18 pKa = 11.84LIATAISFGLFGLGCLGLRR37 pKa = 11.84LVIFPLLSVLPGTAVRR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84EE56 pKa = 3.7RR57 pKa = 11.84ARR59 pKa = 11.84ATVSRR64 pKa = 11.84LFWLFIRR71 pKa = 11.84SMARR75 pKa = 11.84MGVLTYY81 pKa = 10.45DD82 pKa = 3.33VQGAEE87 pKa = 3.81RR88 pKa = 11.84LGRR91 pKa = 11.84PGQMIIANHH100 pKa = 6.84PSLIDD105 pKa = 3.46VVFLIGLVRR114 pKa = 11.84NANCVVKK121 pKa = 10.63QSLWQNPFTRR131 pKa = 11.84SPVRR135 pKa = 11.84ATQYY139 pKa = 10.3ISNDD143 pKa = 2.98GSVDD147 pKa = 3.45MLDD150 pKa = 3.7NAAQALQEE158 pKa = 4.23GQTLIIFPEE167 pKa = 4.83GTRR170 pKa = 11.84TPPGHH175 pKa = 6.15TPAFHH180 pKa = 7.13RR181 pKa = 11.84GAASIALRR189 pKa = 11.84GAKK192 pKa = 9.81VLTPVIIKK200 pKa = 9.17VSPTTLTKK208 pKa = 10.56AEE210 pKa = 3.82PWYY213 pKa = 9.94RR214 pKa = 11.84IPDD217 pKa = 3.56RR218 pKa = 11.84RR219 pKa = 11.84VHH221 pKa = 5.96FSFRR225 pKa = 11.84VGADD229 pKa = 3.13IDD231 pKa = 3.82PHH233 pKa = 5.63TFAALGPPPKK243 pKa = 10.23ASRR246 pKa = 11.84RR247 pKa = 11.84LNDD250 pKa = 3.3FVHH253 pKa = 7.5DD254 pKa = 3.96YY255 pKa = 10.48FIKK258 pKa = 10.57EE259 pKa = 3.91LATDD263 pKa = 3.68EE264 pKa = 4.45RR265 pKa = 11.84SEE267 pKa = 4.14HH268 pKa = 6.52ANN270 pKa = 3.21
MM1 pKa = 8.01DD2 pKa = 6.19LATQPVSTRR11 pKa = 11.84RR12 pKa = 11.84DD13 pKa = 2.99AWCWRR18 pKa = 11.84LIATAISFGLFGLGCLGLRR37 pKa = 11.84LVIFPLLSVLPGTAVRR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84EE56 pKa = 3.7RR57 pKa = 11.84ARR59 pKa = 11.84ATVSRR64 pKa = 11.84LFWLFIRR71 pKa = 11.84SMARR75 pKa = 11.84MGVLTYY81 pKa = 10.45DD82 pKa = 3.33VQGAEE87 pKa = 3.81RR88 pKa = 11.84LGRR91 pKa = 11.84PGQMIIANHH100 pKa = 6.84PSLIDD105 pKa = 3.46VVFLIGLVRR114 pKa = 11.84NANCVVKK121 pKa = 10.63QSLWQNPFTRR131 pKa = 11.84SPVRR135 pKa = 11.84ATQYY139 pKa = 10.3ISNDD143 pKa = 2.98GSVDD147 pKa = 3.45MLDD150 pKa = 3.7NAAQALQEE158 pKa = 4.23GQTLIIFPEE167 pKa = 4.83GTRR170 pKa = 11.84TPPGHH175 pKa = 6.15TPAFHH180 pKa = 7.13RR181 pKa = 11.84GAASIALRR189 pKa = 11.84GAKK192 pKa = 9.81VLTPVIIKK200 pKa = 9.17VSPTTLTKK208 pKa = 10.56AEE210 pKa = 3.82PWYY213 pKa = 9.94RR214 pKa = 11.84IPDD217 pKa = 3.56RR218 pKa = 11.84RR219 pKa = 11.84VHH221 pKa = 5.96FSFRR225 pKa = 11.84VGADD229 pKa = 3.13IDD231 pKa = 3.82PHH233 pKa = 5.63TFAALGPPPKK243 pKa = 10.23ASRR246 pKa = 11.84RR247 pKa = 11.84LNDD250 pKa = 3.3FVHH253 pKa = 7.5DD254 pKa = 3.96YY255 pKa = 10.48FIKK258 pKa = 10.57EE259 pKa = 3.91LATDD263 pKa = 3.68EE264 pKa = 4.45RR265 pKa = 11.84SEE267 pKa = 4.14HH268 pKa = 6.52ANN270 pKa = 3.21
Molecular weight: 30.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1276776 |
29 |
2135 |
321.0 |
35.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.642 ± 0.044 | 1.003 ± 0.012 |
5.38 ± 0.028 | 5.612 ± 0.031 |
3.647 ± 0.024 | 7.701 ± 0.039 |
2.42 ± 0.02 | 4.894 ± 0.027 |
3.759 ± 0.031 | 11.571 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.416 ± 0.019 | 3.2 ± 0.026 |
4.875 ± 0.024 | 4.602 ± 0.031 |
6.141 ± 0.038 | 5.803 ± 0.021 |
5.111 ± 0.03 | 7.178 ± 0.031 |
1.428 ± 0.016 | 2.616 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
