
candidate division MSBL1 archaeon SCGC-AAA382A13
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Euryarchaeota incertae sedis; candidate division MSBL1
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
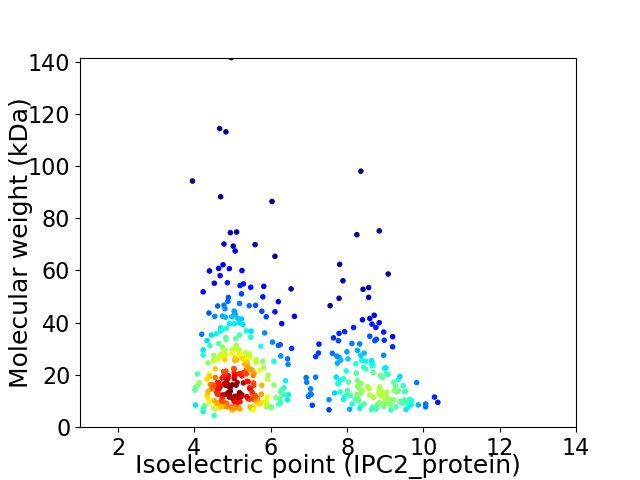
Virtual 2D-PAGE plot for 450 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A133VGW7|A0A133VGW7_9EURY KH type-2 domain-containing protein OS=candidate division MSBL1 archaeon SCGC-AAA382A13 OX=1698279 GN=AKJ50_00175 PE=4 SV=1
MM1 pKa = 7.3ISKK4 pKa = 10.31EE5 pKa = 3.68IFRR8 pKa = 11.84DD9 pKa = 3.53EE10 pKa = 4.42KK11 pKa = 10.98GVSVVLGAILILGMLAAVFPIWYY34 pKa = 8.74NYY36 pKa = 9.11YY37 pKa = 10.29VEE39 pKa = 4.47STMKK43 pKa = 10.58SKK45 pKa = 8.82EE46 pKa = 4.34TNHH49 pKa = 6.17MDD51 pKa = 3.62TVRR54 pKa = 11.84DD55 pKa = 3.69QYY57 pKa = 11.69FEE59 pKa = 4.33LQSKK63 pKa = 8.23VSWMDD68 pKa = 3.33GPEE71 pKa = 3.83TGGKK75 pKa = 9.68VKK77 pKa = 10.48IPMNAGSVRR86 pKa = 11.84FFPGQSPAGTLTIDD100 pKa = 3.67PGSDD104 pKa = 2.5INVEE108 pKa = 3.94NTGVQEE114 pKa = 4.9IITVDD119 pKa = 3.26PGSITFVGQNIYY131 pKa = 10.6FPTQTYY137 pKa = 9.58IFEE140 pKa = 4.85GGHH143 pKa = 5.11VFLVQGEE150 pKa = 4.53AEE152 pKa = 4.92LMASPNEE159 pKa = 3.9EE160 pKa = 4.32LILWGDD166 pKa = 3.9SLEE169 pKa = 4.24VKK171 pKa = 10.01YY172 pKa = 10.36PKK174 pKa = 10.53VVGEE178 pKa = 4.28EE179 pKa = 4.07EE180 pKa = 4.92SISSMVTEE188 pKa = 4.53SVTLAWGGRR197 pKa = 11.84NRR199 pKa = 11.84FPEE202 pKa = 3.89NMEE205 pKa = 3.85WKK207 pKa = 9.98NADD210 pKa = 3.57NIRR213 pKa = 11.84INFEE217 pKa = 3.7TRR219 pKa = 11.84YY220 pKa = 8.44EE221 pKa = 4.25NVWVSYY227 pKa = 11.02LEE229 pKa = 4.07EE230 pKa = 3.91KK231 pKa = 10.95AEE233 pKa = 4.3NIEE236 pKa = 3.94NATIFAEE243 pKa = 4.13ADD245 pKa = 3.23NYY247 pKa = 11.0EE248 pKa = 4.17NGAYY252 pKa = 10.2LQIFNDD258 pKa = 3.6NIRR261 pKa = 11.84YY262 pKa = 7.85LVCATEE268 pKa = 3.95TGEE271 pKa = 4.46DD272 pKa = 3.81VEE274 pKa = 4.81VTNPTVEE281 pKa = 4.03TLEE284 pKa = 4.33NGEE287 pKa = 4.14VWADD291 pKa = 3.21AATLRR296 pKa = 11.84MSYY299 pKa = 10.89DD300 pKa = 3.56FNDD303 pKa = 3.34YY304 pKa = 11.07AYY306 pKa = 10.7GEE308 pKa = 4.47TRR310 pKa = 11.84FKK312 pKa = 9.82WRR314 pKa = 11.84KK315 pKa = 9.27EE316 pKa = 3.64NGTWSATGWSSQYY329 pKa = 11.06GADD332 pKa = 3.82DD333 pKa = 4.22NYY335 pKa = 11.5SEE337 pKa = 5.8LITGIRR343 pKa = 11.84RR344 pKa = 11.84NTTYY348 pKa = 10.49EE349 pKa = 3.66YY350 pKa = 10.71KK351 pKa = 10.85AQIKK355 pKa = 10.02YY356 pKa = 10.3DD357 pKa = 3.82SKK359 pKa = 11.37NKK361 pKa = 9.54EE362 pKa = 4.03GKK364 pKa = 8.34TKK366 pKa = 10.1TFYY369 pKa = 10.35TGNKK373 pKa = 8.29EE374 pKa = 4.21SYY376 pKa = 9.83GGDD379 pKa = 3.29GGRR382 pKa = 11.84INPPSAARR390 pKa = 11.84LVGAEE395 pKa = 4.1KK396 pKa = 10.19TSNDD400 pKa = 2.93TMYY403 pKa = 10.1LTFRR407 pKa = 11.84NTGDD411 pKa = 3.41NTIHH415 pKa = 6.77FGDD418 pKa = 4.22SVVAFYY424 pKa = 10.9KK425 pKa = 10.82EE426 pKa = 4.04EE427 pKa = 4.16IQNKK431 pKa = 8.17TYY433 pKa = 10.75EE434 pKa = 4.44DD435 pKa = 3.63WSSTTLGIGPSPIGGEE451 pKa = 3.92WKK453 pKa = 10.77SPTNQISIPEE463 pKa = 4.09NGEE466 pKa = 3.94TGVSISFSPTDD477 pKa = 3.21AKK479 pKa = 10.68FDD481 pKa = 3.37QGEE484 pKa = 4.15FFVYY488 pKa = 10.16KK489 pKa = 10.8AEE491 pKa = 4.33FSNGTQNIYY500 pKa = 10.2FVNVEE505 pKa = 4.13TTATGNTTPNSPSNPSPSDD524 pKa = 3.18GATLSEE530 pKa = 4.5TTSTTLSVNVSDD542 pKa = 5.61PDD544 pKa = 4.0GDD546 pKa = 4.1SMNVSFYY553 pKa = 10.94DD554 pKa = 3.69ASDD557 pKa = 3.65DD558 pKa = 4.72SIIGTDD564 pKa = 3.17SGVIDD569 pKa = 4.66GGTASVTWDD578 pKa = 3.44SLSAGSSYY586 pKa = 11.48DD587 pKa = 3.07WYY589 pKa = 11.11AVADD593 pKa = 4.79DD594 pKa = 4.75GTDD597 pKa = 3.36STTSSTWSFTVNTAPDD613 pKa = 3.84SPTNPSPSDD622 pKa = 3.51GAVLSSGTSSTTLSVDD638 pKa = 3.07VSDD641 pKa = 5.26PDD643 pKa = 4.16GDD645 pKa = 4.25SMDD648 pKa = 3.33VTFYY652 pKa = 10.3TGEE655 pKa = 4.01GTEE658 pKa = 4.24IGTDD662 pKa = 3.25TGVSSGGTASTTYY675 pKa = 10.86NGLSDD680 pKa = 3.7GNTYY684 pKa = 11.02DD685 pKa = 3.41WYY687 pKa = 11.12AEE689 pKa = 4.03ADD691 pKa = 3.88DD692 pKa = 4.33GHH694 pKa = 7.11DD695 pKa = 3.58VTTSSTYY702 pKa = 10.56SFTIGSVTYY711 pKa = 9.93SYY713 pKa = 10.79TYY715 pKa = 10.82VDD717 pKa = 3.76SEE719 pKa = 4.59TIYY722 pKa = 11.05EE723 pKa = 4.32GGTANFSNAQQDD735 pKa = 3.46DD736 pKa = 4.46GNFEE740 pKa = 4.72NIYY743 pKa = 9.28EE744 pKa = 4.28TDD746 pKa = 3.42QNPQPGQGNIDD757 pKa = 3.35YY758 pKa = 10.68RR759 pKa = 11.84EE760 pKa = 4.05NVQHH764 pKa = 6.79NIVNVSGNNPVFQIEE779 pKa = 4.18YY780 pKa = 9.07MISNDD785 pKa = 3.16TGEE788 pKa = 4.33SVDD791 pKa = 3.76IYY793 pKa = 11.29LEE795 pKa = 4.23NFTSGNWDD803 pKa = 3.65HH804 pKa = 7.16IGQLSSISLDD814 pKa = 3.09NFEE817 pKa = 5.4YY818 pKa = 11.13NMPSDD823 pKa = 4.53YY824 pKa = 10.92ISATDD829 pKa = 4.07NIHH832 pKa = 5.29VRR834 pKa = 11.84YY835 pKa = 9.01VQPTDD840 pKa = 3.32DD841 pKa = 3.91SARR844 pKa = 11.84TDD846 pKa = 4.06LLIDD850 pKa = 3.35YY851 pKa = 9.83CRR853 pKa = 11.84VRR855 pKa = 11.84SEE857 pKa = 4.02TT858 pKa = 3.45
MM1 pKa = 7.3ISKK4 pKa = 10.31EE5 pKa = 3.68IFRR8 pKa = 11.84DD9 pKa = 3.53EE10 pKa = 4.42KK11 pKa = 10.98GVSVVLGAILILGMLAAVFPIWYY34 pKa = 8.74NYY36 pKa = 9.11YY37 pKa = 10.29VEE39 pKa = 4.47STMKK43 pKa = 10.58SKK45 pKa = 8.82EE46 pKa = 4.34TNHH49 pKa = 6.17MDD51 pKa = 3.62TVRR54 pKa = 11.84DD55 pKa = 3.69QYY57 pKa = 11.69FEE59 pKa = 4.33LQSKK63 pKa = 8.23VSWMDD68 pKa = 3.33GPEE71 pKa = 3.83TGGKK75 pKa = 9.68VKK77 pKa = 10.48IPMNAGSVRR86 pKa = 11.84FFPGQSPAGTLTIDD100 pKa = 3.67PGSDD104 pKa = 2.5INVEE108 pKa = 3.94NTGVQEE114 pKa = 4.9IITVDD119 pKa = 3.26PGSITFVGQNIYY131 pKa = 10.6FPTQTYY137 pKa = 9.58IFEE140 pKa = 4.85GGHH143 pKa = 5.11VFLVQGEE150 pKa = 4.53AEE152 pKa = 4.92LMASPNEE159 pKa = 3.9EE160 pKa = 4.32LILWGDD166 pKa = 3.9SLEE169 pKa = 4.24VKK171 pKa = 10.01YY172 pKa = 10.36PKK174 pKa = 10.53VVGEE178 pKa = 4.28EE179 pKa = 4.07EE180 pKa = 4.92SISSMVTEE188 pKa = 4.53SVTLAWGGRR197 pKa = 11.84NRR199 pKa = 11.84FPEE202 pKa = 3.89NMEE205 pKa = 3.85WKK207 pKa = 9.98NADD210 pKa = 3.57NIRR213 pKa = 11.84INFEE217 pKa = 3.7TRR219 pKa = 11.84YY220 pKa = 8.44EE221 pKa = 4.25NVWVSYY227 pKa = 11.02LEE229 pKa = 4.07EE230 pKa = 3.91KK231 pKa = 10.95AEE233 pKa = 4.3NIEE236 pKa = 3.94NATIFAEE243 pKa = 4.13ADD245 pKa = 3.23NYY247 pKa = 11.0EE248 pKa = 4.17NGAYY252 pKa = 10.2LQIFNDD258 pKa = 3.6NIRR261 pKa = 11.84YY262 pKa = 7.85LVCATEE268 pKa = 3.95TGEE271 pKa = 4.46DD272 pKa = 3.81VEE274 pKa = 4.81VTNPTVEE281 pKa = 4.03TLEE284 pKa = 4.33NGEE287 pKa = 4.14VWADD291 pKa = 3.21AATLRR296 pKa = 11.84MSYY299 pKa = 10.89DD300 pKa = 3.56FNDD303 pKa = 3.34YY304 pKa = 11.07AYY306 pKa = 10.7GEE308 pKa = 4.47TRR310 pKa = 11.84FKK312 pKa = 9.82WRR314 pKa = 11.84KK315 pKa = 9.27EE316 pKa = 3.64NGTWSATGWSSQYY329 pKa = 11.06GADD332 pKa = 3.82DD333 pKa = 4.22NYY335 pKa = 11.5SEE337 pKa = 5.8LITGIRR343 pKa = 11.84RR344 pKa = 11.84NTTYY348 pKa = 10.49EE349 pKa = 3.66YY350 pKa = 10.71KK351 pKa = 10.85AQIKK355 pKa = 10.02YY356 pKa = 10.3DD357 pKa = 3.82SKK359 pKa = 11.37NKK361 pKa = 9.54EE362 pKa = 4.03GKK364 pKa = 8.34TKK366 pKa = 10.1TFYY369 pKa = 10.35TGNKK373 pKa = 8.29EE374 pKa = 4.21SYY376 pKa = 9.83GGDD379 pKa = 3.29GGRR382 pKa = 11.84INPPSAARR390 pKa = 11.84LVGAEE395 pKa = 4.1KK396 pKa = 10.19TSNDD400 pKa = 2.93TMYY403 pKa = 10.1LTFRR407 pKa = 11.84NTGDD411 pKa = 3.41NTIHH415 pKa = 6.77FGDD418 pKa = 4.22SVVAFYY424 pKa = 10.9KK425 pKa = 10.82EE426 pKa = 4.04EE427 pKa = 4.16IQNKK431 pKa = 8.17TYY433 pKa = 10.75EE434 pKa = 4.44DD435 pKa = 3.63WSSTTLGIGPSPIGGEE451 pKa = 3.92WKK453 pKa = 10.77SPTNQISIPEE463 pKa = 4.09NGEE466 pKa = 3.94TGVSISFSPTDD477 pKa = 3.21AKK479 pKa = 10.68FDD481 pKa = 3.37QGEE484 pKa = 4.15FFVYY488 pKa = 10.16KK489 pKa = 10.8AEE491 pKa = 4.33FSNGTQNIYY500 pKa = 10.2FVNVEE505 pKa = 4.13TTATGNTTPNSPSNPSPSDD524 pKa = 3.18GATLSEE530 pKa = 4.5TTSTTLSVNVSDD542 pKa = 5.61PDD544 pKa = 4.0GDD546 pKa = 4.1SMNVSFYY553 pKa = 10.94DD554 pKa = 3.69ASDD557 pKa = 3.65DD558 pKa = 4.72SIIGTDD564 pKa = 3.17SGVIDD569 pKa = 4.66GGTASVTWDD578 pKa = 3.44SLSAGSSYY586 pKa = 11.48DD587 pKa = 3.07WYY589 pKa = 11.11AVADD593 pKa = 4.79DD594 pKa = 4.75GTDD597 pKa = 3.36STTSSTWSFTVNTAPDD613 pKa = 3.84SPTNPSPSDD622 pKa = 3.51GAVLSSGTSSTTLSVDD638 pKa = 3.07VSDD641 pKa = 5.26PDD643 pKa = 4.16GDD645 pKa = 4.25SMDD648 pKa = 3.33VTFYY652 pKa = 10.3TGEE655 pKa = 4.01GTEE658 pKa = 4.24IGTDD662 pKa = 3.25TGVSSGGTASTTYY675 pKa = 10.86NGLSDD680 pKa = 3.7GNTYY684 pKa = 11.02DD685 pKa = 3.41WYY687 pKa = 11.12AEE689 pKa = 4.03ADD691 pKa = 3.88DD692 pKa = 4.33GHH694 pKa = 7.11DD695 pKa = 3.58VTTSSTYY702 pKa = 10.56SFTIGSVTYY711 pKa = 9.93SYY713 pKa = 10.79TYY715 pKa = 10.82VDD717 pKa = 3.76SEE719 pKa = 4.59TIYY722 pKa = 11.05EE723 pKa = 4.32GGTANFSNAQQDD735 pKa = 3.46DD736 pKa = 4.46GNFEE740 pKa = 4.72NIYY743 pKa = 9.28EE744 pKa = 4.28TDD746 pKa = 3.42QNPQPGQGNIDD757 pKa = 3.35YY758 pKa = 10.68RR759 pKa = 11.84EE760 pKa = 4.05NVQHH764 pKa = 6.79NIVNVSGNNPVFQIEE779 pKa = 4.18YY780 pKa = 9.07MISNDD785 pKa = 3.16TGEE788 pKa = 4.33SVDD791 pKa = 3.76IYY793 pKa = 11.29LEE795 pKa = 4.23NFTSGNWDD803 pKa = 3.65HH804 pKa = 7.16IGQLSSISLDD814 pKa = 3.09NFEE817 pKa = 5.4YY818 pKa = 11.13NMPSDD823 pKa = 4.53YY824 pKa = 10.92ISATDD829 pKa = 4.07NIHH832 pKa = 5.29VRR834 pKa = 11.84YY835 pKa = 9.01VQPTDD840 pKa = 3.32DD841 pKa = 3.91SARR844 pKa = 11.84TDD846 pKa = 4.06LLIDD850 pKa = 3.35YY851 pKa = 9.83CRR853 pKa = 11.84VRR855 pKa = 11.84SEE857 pKa = 4.02TT858 pKa = 3.45
Molecular weight: 94.24 kDa
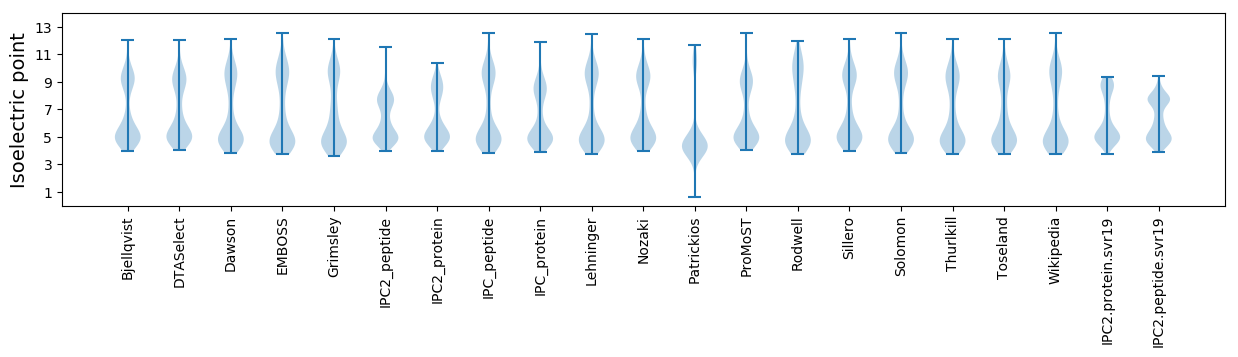
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A133VFX7|A0A133VFX7_9EURY Uncharacterized protein OS=candidate division MSBL1 archaeon SCGC-AAA382A13 OX=1698279 GN=AKJ50_01195 PE=4 SV=1
RR1 pKa = 7.89DD2 pKa = 3.49NCDD5 pKa = 3.09LVVDD9 pKa = 3.91VGGIPRR15 pKa = 11.84FYY17 pKa = 9.46PKK19 pKa = 10.38KK20 pKa = 10.66GITLKK25 pKa = 10.87RR26 pKa = 11.84KK27 pKa = 9.82GSWSWTEE34 pKa = 3.48MLMNLIEE41 pKa = 4.93NPQGWLRR48 pKa = 11.84DD49 pKa = 3.39YY50 pKa = 10.57HH51 pKa = 5.96QRR53 pKa = 11.84SNIEE57 pKa = 4.21TIYY60 pKa = 10.0STLKK64 pKa = 10.27RR65 pKa = 11.84DD66 pKa = 3.45FQTRR70 pKa = 11.84PRR72 pKa = 11.84SRR74 pKa = 11.84NLNRR78 pKa = 11.84RR79 pKa = 11.84GG80 pKa = 3.29
RR1 pKa = 7.89DD2 pKa = 3.49NCDD5 pKa = 3.09LVVDD9 pKa = 3.91VGGIPRR15 pKa = 11.84FYY17 pKa = 9.46PKK19 pKa = 10.38KK20 pKa = 10.66GITLKK25 pKa = 10.87RR26 pKa = 11.84KK27 pKa = 9.82GSWSWTEE34 pKa = 3.48MLMNLIEE41 pKa = 4.93NPQGWLRR48 pKa = 11.84DD49 pKa = 3.39YY50 pKa = 10.57HH51 pKa = 5.96QRR53 pKa = 11.84SNIEE57 pKa = 4.21TIYY60 pKa = 10.0STLKK64 pKa = 10.27RR65 pKa = 11.84DD66 pKa = 3.45FQTRR70 pKa = 11.84PRR72 pKa = 11.84SRR74 pKa = 11.84NLNRR78 pKa = 11.84RR79 pKa = 11.84GG80 pKa = 3.29
Molecular weight: 9.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
96092 |
41 |
1206 |
213.5 |
24.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.969 ± 0.131 | 1.0 ± 0.052 |
5.749 ± 0.112 | 9.935 ± 0.158 |
3.864 ± 0.101 | 6.94 ± 0.146 |
1.624 ± 0.048 | 7.599 ± 0.125 |
8.473 ± 0.188 | 9.037 ± 0.139 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.28 ± 0.056 | 4.241 ± 0.08 |
3.948 ± 0.061 | 2.414 ± 0.055 |
5.052 ± 0.094 | 6.383 ± 0.106 |
4.907 ± 0.089 | 6.457 ± 0.116 |
1.099 ± 0.045 | 3.026 ± 0.081 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
