
Porcine torque teno virus 2
Taxonomy: Viruses; Anelloviridae; Iotatorquevirus; Torque teno sus virus 1b
Average proteome isoelectric point is 7.07
Get precalculated fractions of proteins
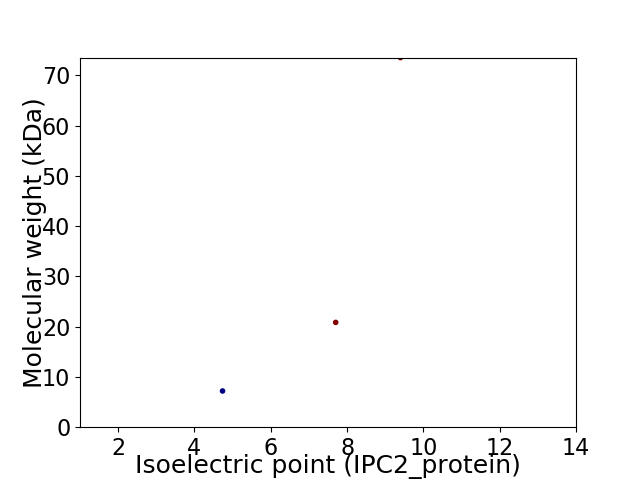
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|G8Z9V8|G8Z9V8_9VIRU Capsid protein OS=Porcine torque teno virus 2 OX=871765 PE=3 SV=1
MM1 pKa = 7.51EE2 pKa = 4.92EE3 pKa = 3.49RR4 pKa = 11.84WLTVAYY10 pKa = 8.84CAHH13 pKa = 6.88GLFCGCKK20 pKa = 9.81DD21 pKa = 3.68PKK23 pKa = 10.8KK24 pKa = 10.09HH25 pKa = 6.71LEE27 pKa = 3.92KK28 pKa = 10.88CLTDD32 pKa = 5.18AIADD36 pKa = 3.85AEE38 pKa = 4.49GDD40 pKa = 3.39RR41 pKa = 11.84HH42 pKa = 6.31EE43 pKa = 5.77DD44 pKa = 3.32GGTGGGDD51 pKa = 2.92ATFDD55 pKa = 3.25IGIDD59 pKa = 3.51ALLAAAAQRR68 pKa = 3.68
MM1 pKa = 7.51EE2 pKa = 4.92EE3 pKa = 3.49RR4 pKa = 11.84WLTVAYY10 pKa = 8.84CAHH13 pKa = 6.88GLFCGCKK20 pKa = 9.81DD21 pKa = 3.68PKK23 pKa = 10.8KK24 pKa = 10.09HH25 pKa = 6.71LEE27 pKa = 3.92KK28 pKa = 10.88CLTDD32 pKa = 5.18AIADD36 pKa = 3.85AEE38 pKa = 4.49GDD40 pKa = 3.39RR41 pKa = 11.84HH42 pKa = 6.31EE43 pKa = 5.77DD44 pKa = 3.32GGTGGGDD51 pKa = 2.92ATFDD55 pKa = 3.25IGIDD59 pKa = 3.51ALLAAAAQRR68 pKa = 3.68
Molecular weight: 7.21 kDa
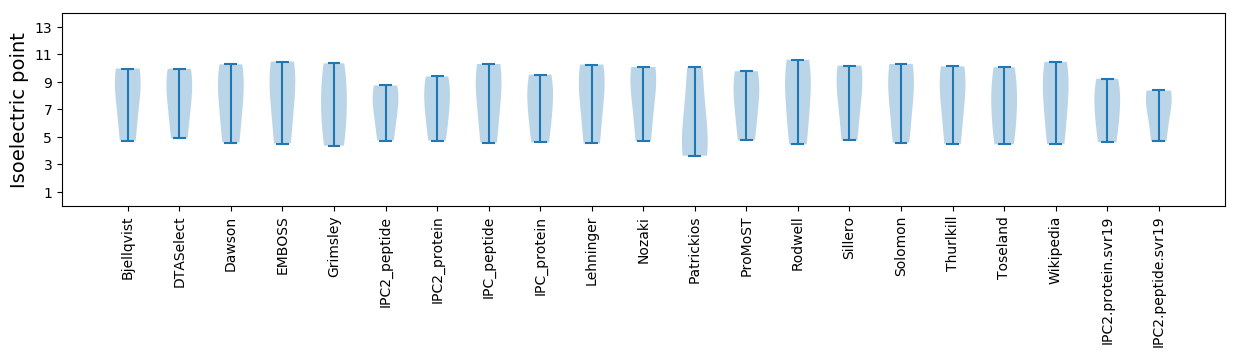
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G8Z9V8|G8Z9V8_9VIRU Capsid protein OS=Porcine torque teno virus 2 OX=871765 PE=3 SV=1
MM1 pKa = 8.01PYY3 pKa = 9.82RR4 pKa = 11.84RR5 pKa = 11.84YY6 pKa = 9.5RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84PTRR15 pKa = 11.84RR16 pKa = 11.84WRR18 pKa = 11.84HH19 pKa = 4.47RR20 pKa = 11.84RR21 pKa = 11.84WRR23 pKa = 11.84RR24 pKa = 11.84YY25 pKa = 7.81FRR27 pKa = 11.84YY28 pKa = 9.49RR29 pKa = 11.84YY30 pKa = 9.15RR31 pKa = 11.84RR32 pKa = 11.84APRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84TKK40 pKa = 8.79VRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84KK47 pKa = 9.72APVIQWFPPSRR58 pKa = 11.84RR59 pKa = 11.84TCLIEE64 pKa = 4.6GFWPLSHH71 pKa = 6.02GHH73 pKa = 6.06WFRR76 pKa = 11.84TCPPMRR82 pKa = 11.84RR83 pKa = 11.84LNGLIFTGGGCDD95 pKa = 2.95WTQWSLQNLYY105 pKa = 10.4HH106 pKa = 7.36EE107 pKa = 4.67KK108 pKa = 10.89LNWRR112 pKa = 11.84NIWTASNVGMEE123 pKa = 4.49FARR126 pKa = 11.84FLRR129 pKa = 11.84GKK131 pKa = 10.07FYY133 pKa = 10.73FFRR136 pKa = 11.84HH137 pKa = 5.6PWRR140 pKa = 11.84SYY142 pKa = 10.33IVTWDD147 pKa = 3.16QDD149 pKa = 3.94IPCKK153 pKa = 9.77PLPYY157 pKa = 10.21QNLQPLLMLLKK168 pKa = 9.96KK169 pKa = 8.04QHH171 pKa = 6.63KK172 pKa = 9.82LVLSQKK178 pKa = 10.3DD179 pKa = 3.66CNPNRR184 pKa = 11.84KK185 pKa = 8.65QKK187 pKa = 10.48PVTLKK192 pKa = 10.25FRR194 pKa = 11.84PPPKK198 pKa = 9.71LTSQWRR204 pKa = 11.84LSRR207 pKa = 11.84EE208 pKa = 3.69LSKK211 pKa = 10.89IPLIRR216 pKa = 11.84LGISLIDD223 pKa = 4.34LSEE226 pKa = 4.04PWLEE230 pKa = 3.84GWGNAFYY237 pKa = 11.12SVLGYY242 pKa = 9.35EE243 pKa = 4.41ASKK246 pKa = 11.09HH247 pKa = 4.81SGRR250 pKa = 11.84WSNWTQMKK258 pKa = 9.28YY259 pKa = 9.31FWIYY263 pKa = 9.61DD264 pKa = 3.69TGVGNAVYY272 pKa = 10.59VILLKK277 pKa = 10.49KK278 pKa = 10.43DD279 pKa = 3.4VSDD282 pKa = 4.35NPGDD286 pKa = 3.61MATQFVTGSGQHH298 pKa = 6.95PDD300 pKa = 4.02AIDD303 pKa = 4.09HH304 pKa = 6.49IEE306 pKa = 4.08MVNEE310 pKa = 4.58GWPYY314 pKa = 9.43WLFFYY319 pKa = 10.29GQSEE323 pKa = 4.1QGIKK327 pKa = 10.27KK328 pKa = 9.44LAHH331 pKa = 6.39DD332 pKa = 3.59QDD334 pKa = 3.53IVRR337 pKa = 11.84EE338 pKa = 4.03YY339 pKa = 11.48ARR341 pKa = 11.84DD342 pKa = 3.74PKK344 pKa = 10.9SKK346 pKa = 10.0KK347 pKa = 10.37LKK349 pKa = 10.0IGVIGWASSNYY360 pKa = 6.33TTAGSNQNTQSQTPEE375 pKa = 4.64AIQGGYY381 pKa = 8.0VAYY384 pKa = 10.03AGSRR388 pKa = 11.84IPGAGSITNLFQMGWPGDD406 pKa = 4.08QNWPPTNQDD415 pKa = 2.76QTNFNWGLRR424 pKa = 11.84GLCVLRR430 pKa = 11.84DD431 pKa = 3.75NMKK434 pKa = 10.72LGAQEE439 pKa = 5.52LDD441 pKa = 4.35DD442 pKa = 4.54EE443 pKa = 4.78CTMLSLFGPFVEE455 pKa = 4.88KK456 pKa = 11.02ANTAFATNDD465 pKa = 3.26PKK467 pKa = 11.25YY468 pKa = 10.58FRR470 pKa = 11.84PEE472 pKa = 3.71LKK474 pKa = 10.24DD475 pKa = 3.49YY476 pKa = 11.59NVVTKK481 pKa = 10.43YY482 pKa = 10.82AFKK485 pKa = 10.32FQWGGHH491 pKa = 3.76GTEE494 pKa = 4.41RR495 pKa = 11.84FKK497 pKa = 9.27TTIGDD502 pKa = 3.85PSTIPCPFEE511 pKa = 4.92PGEE514 pKa = 4.27RR515 pKa = 11.84YY516 pKa = 8.96HH517 pKa = 8.27HH518 pKa = 6.43GVQDD522 pKa = 3.65PAKK525 pKa = 10.05VQNTVLNPWDD535 pKa = 4.05YY536 pKa = 11.7DD537 pKa = 3.35CDD539 pKa = 4.96GIVRR543 pKa = 11.84TDD545 pKa = 3.21TLKK548 pKa = 10.96RR549 pKa = 11.84LLEE552 pKa = 4.48LPTEE556 pKa = 4.27TEE558 pKa = 4.35EE559 pKa = 4.41TEE561 pKa = 3.86KK562 pKa = 10.75AYY564 pKa = 9.86PLLGQKK570 pKa = 7.42TEE572 pKa = 4.39KK573 pKa = 10.65EE574 pKa = 4.16PLSDD578 pKa = 3.53SDD580 pKa = 3.95EE581 pKa = 4.33EE582 pKa = 4.55SVISSTSSGSSQEE595 pKa = 4.36EE596 pKa = 3.9EE597 pKa = 4.05TQRR600 pKa = 11.84RR601 pKa = 11.84RR602 pKa = 11.84QHH604 pKa = 6.02KK605 pKa = 8.05PSKK608 pKa = 10.13RR609 pKa = 11.84RR610 pKa = 11.84LLKK613 pKa = 10.16HH614 pKa = 5.57LQRR617 pKa = 11.84VVKK620 pKa = 10.41RR621 pKa = 11.84MKK623 pKa = 9.97TLL625 pKa = 3.31
MM1 pKa = 8.01PYY3 pKa = 9.82RR4 pKa = 11.84RR5 pKa = 11.84YY6 pKa = 9.5RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84PTRR15 pKa = 11.84RR16 pKa = 11.84WRR18 pKa = 11.84HH19 pKa = 4.47RR20 pKa = 11.84RR21 pKa = 11.84WRR23 pKa = 11.84RR24 pKa = 11.84YY25 pKa = 7.81FRR27 pKa = 11.84YY28 pKa = 9.49RR29 pKa = 11.84YY30 pKa = 9.15RR31 pKa = 11.84RR32 pKa = 11.84APRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84TKK40 pKa = 8.79VRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84KK47 pKa = 9.72APVIQWFPPSRR58 pKa = 11.84RR59 pKa = 11.84TCLIEE64 pKa = 4.6GFWPLSHH71 pKa = 6.02GHH73 pKa = 6.06WFRR76 pKa = 11.84TCPPMRR82 pKa = 11.84RR83 pKa = 11.84LNGLIFTGGGCDD95 pKa = 2.95WTQWSLQNLYY105 pKa = 10.4HH106 pKa = 7.36EE107 pKa = 4.67KK108 pKa = 10.89LNWRR112 pKa = 11.84NIWTASNVGMEE123 pKa = 4.49FARR126 pKa = 11.84FLRR129 pKa = 11.84GKK131 pKa = 10.07FYY133 pKa = 10.73FFRR136 pKa = 11.84HH137 pKa = 5.6PWRR140 pKa = 11.84SYY142 pKa = 10.33IVTWDD147 pKa = 3.16QDD149 pKa = 3.94IPCKK153 pKa = 9.77PLPYY157 pKa = 10.21QNLQPLLMLLKK168 pKa = 9.96KK169 pKa = 8.04QHH171 pKa = 6.63KK172 pKa = 9.82LVLSQKK178 pKa = 10.3DD179 pKa = 3.66CNPNRR184 pKa = 11.84KK185 pKa = 8.65QKK187 pKa = 10.48PVTLKK192 pKa = 10.25FRR194 pKa = 11.84PPPKK198 pKa = 9.71LTSQWRR204 pKa = 11.84LSRR207 pKa = 11.84EE208 pKa = 3.69LSKK211 pKa = 10.89IPLIRR216 pKa = 11.84LGISLIDD223 pKa = 4.34LSEE226 pKa = 4.04PWLEE230 pKa = 3.84GWGNAFYY237 pKa = 11.12SVLGYY242 pKa = 9.35EE243 pKa = 4.41ASKK246 pKa = 11.09HH247 pKa = 4.81SGRR250 pKa = 11.84WSNWTQMKK258 pKa = 9.28YY259 pKa = 9.31FWIYY263 pKa = 9.61DD264 pKa = 3.69TGVGNAVYY272 pKa = 10.59VILLKK277 pKa = 10.49KK278 pKa = 10.43DD279 pKa = 3.4VSDD282 pKa = 4.35NPGDD286 pKa = 3.61MATQFVTGSGQHH298 pKa = 6.95PDD300 pKa = 4.02AIDD303 pKa = 4.09HH304 pKa = 6.49IEE306 pKa = 4.08MVNEE310 pKa = 4.58GWPYY314 pKa = 9.43WLFFYY319 pKa = 10.29GQSEE323 pKa = 4.1QGIKK327 pKa = 10.27KK328 pKa = 9.44LAHH331 pKa = 6.39DD332 pKa = 3.59QDD334 pKa = 3.53IVRR337 pKa = 11.84EE338 pKa = 4.03YY339 pKa = 11.48ARR341 pKa = 11.84DD342 pKa = 3.74PKK344 pKa = 10.9SKK346 pKa = 10.0KK347 pKa = 10.37LKK349 pKa = 10.0IGVIGWASSNYY360 pKa = 6.33TTAGSNQNTQSQTPEE375 pKa = 4.64AIQGGYY381 pKa = 8.0VAYY384 pKa = 10.03AGSRR388 pKa = 11.84IPGAGSITNLFQMGWPGDD406 pKa = 4.08QNWPPTNQDD415 pKa = 2.76QTNFNWGLRR424 pKa = 11.84GLCVLRR430 pKa = 11.84DD431 pKa = 3.75NMKK434 pKa = 10.72LGAQEE439 pKa = 5.52LDD441 pKa = 4.35DD442 pKa = 4.54EE443 pKa = 4.78CTMLSLFGPFVEE455 pKa = 4.88KK456 pKa = 11.02ANTAFATNDD465 pKa = 3.26PKK467 pKa = 11.25YY468 pKa = 10.58FRR470 pKa = 11.84PEE472 pKa = 3.71LKK474 pKa = 10.24DD475 pKa = 3.49YY476 pKa = 11.59NVVTKK481 pKa = 10.43YY482 pKa = 10.82AFKK485 pKa = 10.32FQWGGHH491 pKa = 3.76GTEE494 pKa = 4.41RR495 pKa = 11.84FKK497 pKa = 9.27TTIGDD502 pKa = 3.85PSTIPCPFEE511 pKa = 4.92PGEE514 pKa = 4.27RR515 pKa = 11.84YY516 pKa = 8.96HH517 pKa = 8.27HH518 pKa = 6.43GVQDD522 pKa = 3.65PAKK525 pKa = 10.05VQNTVLNPWDD535 pKa = 4.05YY536 pKa = 11.7DD537 pKa = 3.35CDD539 pKa = 4.96GIVRR543 pKa = 11.84TDD545 pKa = 3.21TLKK548 pKa = 10.96RR549 pKa = 11.84LLEE552 pKa = 4.48LPTEE556 pKa = 4.27TEE558 pKa = 4.35EE559 pKa = 4.41TEE561 pKa = 3.86KK562 pKa = 10.75AYY564 pKa = 9.86PLLGQKK570 pKa = 7.42TEE572 pKa = 4.39KK573 pKa = 10.65EE574 pKa = 4.16PLSDD578 pKa = 3.53SDD580 pKa = 3.95EE581 pKa = 4.33EE582 pKa = 4.55SVISSTSSGSSQEE595 pKa = 4.36EE596 pKa = 3.9EE597 pKa = 4.05TQRR600 pKa = 11.84RR601 pKa = 11.84RR602 pKa = 11.84QHH604 pKa = 6.02KK605 pKa = 8.05PSKK608 pKa = 10.13RR609 pKa = 11.84RR610 pKa = 11.84LLKK613 pKa = 10.16HH614 pKa = 5.57LQRR617 pKa = 11.84VVKK620 pKa = 10.41RR621 pKa = 11.84MKK623 pKa = 9.97TLL625 pKa = 3.31
Molecular weight: 73.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
879 |
68 |
625 |
293.0 |
33.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.371 ± 3.633 | 1.934 ± 1.038 |
5.802 ± 1.854 | 5.688 ± 0.96 |
3.641 ± 0.678 | 7.85 ± 1.494 |
2.73 ± 0.525 | 4.323 ± 0.433 |
6.94 ± 0.645 | 8.191 ± 0.269 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.706 ± 0.08 | 3.982 ± 0.986 |
5.802 ± 1.564 | 4.778 ± 0.921 |
8.987 ± 1.328 | 4.664 ± 1.782 |
6.143 ± 0.129 | 3.641 ± 0.932 |
3.299 ± 0.934 | 3.527 ± 0.872 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
