
Acetatifactor muris
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Acetatifactor
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
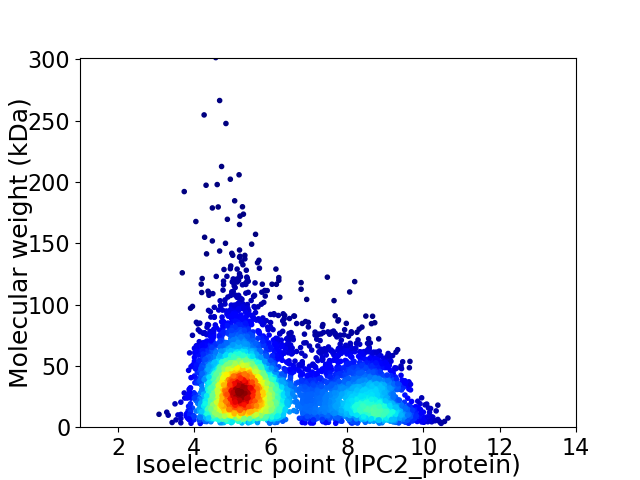
Virtual 2D-PAGE plot for 5687 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2K4ZN03|A0A2K4ZN03_9FIRM Prophage phiRv2 integrase OS=Acetatifactor muris OX=879566 GN=AMURIS_04569 PE=4 SV=1
MM1 pKa = 7.62KK2 pKa = 10.66KK3 pKa = 7.68MTALFLSLALVFGCVGCGNDD23 pKa = 4.44DD24 pKa = 3.84GQASGSSDD32 pKa = 3.09SSGTSVEE39 pKa = 4.35SSDD42 pKa = 4.88ASDD45 pKa = 3.83ASTEE49 pKa = 4.07SSTEE53 pKa = 3.89STAEE57 pKa = 3.86AQEE60 pKa = 4.4SSSEE64 pKa = 3.99ASDD67 pKa = 3.31AGEE70 pKa = 4.14DD71 pKa = 3.72VKK73 pKa = 11.44SEE75 pKa = 3.67GVMTYY80 pKa = 10.36EE81 pKa = 4.51EE82 pKa = 4.28YY83 pKa = 10.93AAAALDD89 pKa = 3.83TEE91 pKa = 4.87VVIEE95 pKa = 4.92AYY97 pKa = 10.34VQAKK101 pKa = 8.46QSWWEE106 pKa = 3.75KK107 pKa = 10.83DD108 pKa = 3.37GQGCCTVYY116 pKa = 10.94AQDD119 pKa = 3.42KK120 pKa = 10.36DD121 pKa = 3.39GGYY124 pKa = 10.46FLYY127 pKa = 10.74EE128 pKa = 4.06MACSEE133 pKa = 4.07EE134 pKa = 5.39DD135 pKa = 3.66YY136 pKa = 11.52AKK138 pKa = 10.58LVPGTKK144 pKa = 9.29IKK146 pKa = 9.86VTGFKK151 pKa = 10.81AEE153 pKa = 3.69WSGEE157 pKa = 3.88FEE159 pKa = 4.8IIDD162 pKa = 3.4AAFEE166 pKa = 4.16IGEE169 pKa = 4.13GSYY172 pKa = 10.1IAEE175 pKa = 4.53AEE177 pKa = 4.32DD178 pKa = 3.46VTALLGQDD186 pKa = 4.53DD187 pKa = 5.1LVNHH191 pKa = 5.58QNKK194 pKa = 8.7FVSFKK199 pKa = 11.12GMTVEE204 pKa = 4.33AAKK207 pKa = 10.72DD208 pKa = 3.46PEE210 pKa = 4.24GNIIKK215 pKa = 10.6DD216 pKa = 3.47EE217 pKa = 4.71AGNEE221 pKa = 3.85AAFLYY226 pKa = 10.39NWDD229 pKa = 3.9GSGQDD234 pKa = 4.14GDD236 pKa = 4.73DD237 pKa = 3.72LYY239 pKa = 11.84FNVSVNGATYY249 pKa = 8.66TFTVEE254 pKa = 5.73SYY256 pKa = 11.11LCDD259 pKa = 3.35STTDD263 pKa = 2.74VYY265 pKa = 11.6AAVKK269 pKa = 9.49GLKK272 pKa = 10.33VGDD275 pKa = 4.34KK276 pKa = 10.2IDD278 pKa = 3.83MEE280 pKa = 4.68GFLYY284 pKa = 9.77WYY286 pKa = 10.36NGVNPHH292 pKa = 5.61ITSVKK297 pKa = 9.83AANN300 pKa = 3.38
MM1 pKa = 7.62KK2 pKa = 10.66KK3 pKa = 7.68MTALFLSLALVFGCVGCGNDD23 pKa = 4.44DD24 pKa = 3.84GQASGSSDD32 pKa = 3.09SSGTSVEE39 pKa = 4.35SSDD42 pKa = 4.88ASDD45 pKa = 3.83ASTEE49 pKa = 4.07SSTEE53 pKa = 3.89STAEE57 pKa = 3.86AQEE60 pKa = 4.4SSSEE64 pKa = 3.99ASDD67 pKa = 3.31AGEE70 pKa = 4.14DD71 pKa = 3.72VKK73 pKa = 11.44SEE75 pKa = 3.67GVMTYY80 pKa = 10.36EE81 pKa = 4.51EE82 pKa = 4.28YY83 pKa = 10.93AAAALDD89 pKa = 3.83TEE91 pKa = 4.87VVIEE95 pKa = 4.92AYY97 pKa = 10.34VQAKK101 pKa = 8.46QSWWEE106 pKa = 3.75KK107 pKa = 10.83DD108 pKa = 3.37GQGCCTVYY116 pKa = 10.94AQDD119 pKa = 3.42KK120 pKa = 10.36DD121 pKa = 3.39GGYY124 pKa = 10.46FLYY127 pKa = 10.74EE128 pKa = 4.06MACSEE133 pKa = 4.07EE134 pKa = 5.39DD135 pKa = 3.66YY136 pKa = 11.52AKK138 pKa = 10.58LVPGTKK144 pKa = 9.29IKK146 pKa = 9.86VTGFKK151 pKa = 10.81AEE153 pKa = 3.69WSGEE157 pKa = 3.88FEE159 pKa = 4.8IIDD162 pKa = 3.4AAFEE166 pKa = 4.16IGEE169 pKa = 4.13GSYY172 pKa = 10.1IAEE175 pKa = 4.53AEE177 pKa = 4.32DD178 pKa = 3.46VTALLGQDD186 pKa = 4.53DD187 pKa = 5.1LVNHH191 pKa = 5.58QNKK194 pKa = 8.7FVSFKK199 pKa = 11.12GMTVEE204 pKa = 4.33AAKK207 pKa = 10.72DD208 pKa = 3.46PEE210 pKa = 4.24GNIIKK215 pKa = 10.6DD216 pKa = 3.47EE217 pKa = 4.71AGNEE221 pKa = 3.85AAFLYY226 pKa = 10.39NWDD229 pKa = 3.9GSGQDD234 pKa = 4.14GDD236 pKa = 4.73DD237 pKa = 3.72LYY239 pKa = 11.84FNVSVNGATYY249 pKa = 8.66TFTVEE254 pKa = 5.73SYY256 pKa = 11.11LCDD259 pKa = 3.35STTDD263 pKa = 2.74VYY265 pKa = 11.6AAVKK269 pKa = 9.49GLKK272 pKa = 10.33VGDD275 pKa = 4.34KK276 pKa = 10.2IDD278 pKa = 3.83MEE280 pKa = 4.68GFLYY284 pKa = 9.77WYY286 pKa = 10.36NGVNPHH292 pKa = 5.61ITSVKK297 pKa = 9.83AANN300 pKa = 3.38
Molecular weight: 32.11 kDa
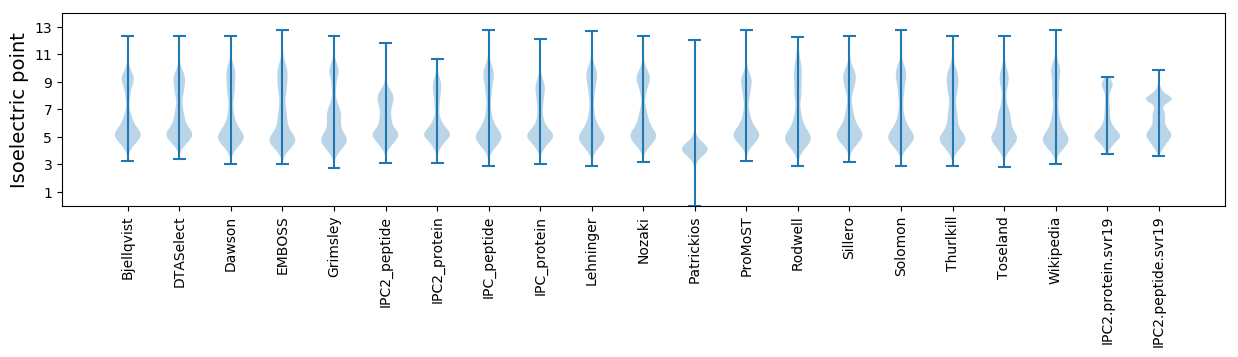
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2K4ZDR9|A0A2K4ZDR9_9FIRM 3-isopropylmalate dehydratase large subunit OS=Acetatifactor muris OX=879566 GN=dmdA_2 PE=3 SV=1
MM1 pKa = 7.68KK2 pKa = 10.19NRR4 pKa = 11.84EE5 pKa = 4.53KK6 pKa = 10.31IRR8 pKa = 11.84EE9 pKa = 4.08MTTTALLAALLAVMGMFRR27 pKa = 11.84LPSILPGCEE36 pKa = 3.88FQLSAPFAVCLAACFGFRR54 pKa = 11.84RR55 pKa = 11.84YY56 pKa = 9.88FKK58 pKa = 10.47IGVLASMMNLILGTHH73 pKa = 6.62TIVNVTIAMIFRR85 pKa = 11.84LVAGGILSLFGVNPVTIAVSGPLGTVAGRR114 pKa = 11.84VVLGAISDD122 pKa = 4.19TNPLALTAAAAPGMVFTAVGASVMYY147 pKa = 9.64PVMRR151 pKa = 11.84HH152 pKa = 4.51LVRR155 pKa = 11.84RR156 pKa = 11.84EE157 pKa = 3.6KK158 pKa = 10.97NAA160 pKa = 3.36
MM1 pKa = 7.68KK2 pKa = 10.19NRR4 pKa = 11.84EE5 pKa = 4.53KK6 pKa = 10.31IRR8 pKa = 11.84EE9 pKa = 4.08MTTTALLAALLAVMGMFRR27 pKa = 11.84LPSILPGCEE36 pKa = 3.88FQLSAPFAVCLAACFGFRR54 pKa = 11.84RR55 pKa = 11.84YY56 pKa = 9.88FKK58 pKa = 10.47IGVLASMMNLILGTHH73 pKa = 6.62TIVNVTIAMIFRR85 pKa = 11.84LVAGGILSLFGVNPVTIAVSGPLGTVAGRR114 pKa = 11.84VVLGAISDD122 pKa = 4.19TNPLALTAAAAPGMVFTAVGASVMYY147 pKa = 9.64PVMRR151 pKa = 11.84HH152 pKa = 4.51LVRR155 pKa = 11.84RR156 pKa = 11.84EE157 pKa = 3.6KK158 pKa = 10.97NAA160 pKa = 3.36
Molecular weight: 16.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1754436 |
29 |
2719 |
308.5 |
34.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.624 ± 0.032 | 1.612 ± 0.015 |
5.455 ± 0.028 | 8.128 ± 0.038 |
4.103 ± 0.026 | 7.309 ± 0.033 |
1.718 ± 0.015 | 6.606 ± 0.03 |
6.066 ± 0.035 | 9.151 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.055 ± 0.016 | 4.145 ± 0.025 |
3.377 ± 0.022 | 3.375 ± 0.019 |
5.457 ± 0.029 | 5.706 ± 0.038 |
5.143 ± 0.023 | 6.525 ± 0.028 |
1.135 ± 0.014 | 4.309 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
