
Capybara microvirus Cap1_SP_216
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.37
Get precalculated fractions of proteins
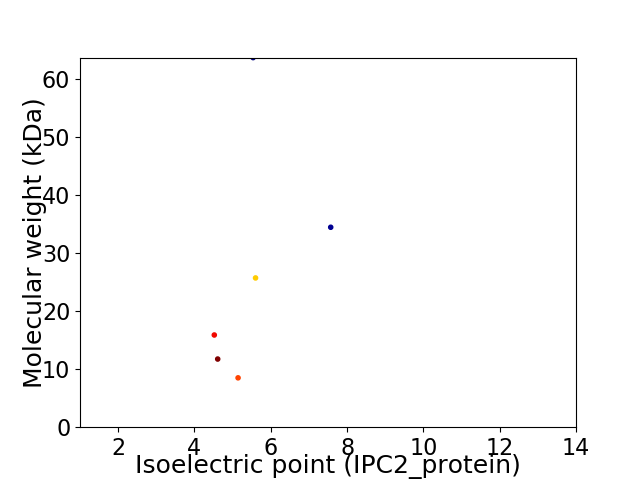
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W4M2|A0A4P8W4M2_9VIRU Replication initiator protein OS=Capybara microvirus Cap1_SP_216 OX=2585411 PE=4 SV=1
MM1 pKa = 7.42KK2 pKa = 10.33KK3 pKa = 10.42LYY5 pKa = 9.56TIHH8 pKa = 7.64DD9 pKa = 4.63KK10 pKa = 10.77IANKK14 pKa = 10.5CGDD17 pKa = 3.32IFTAEE22 pKa = 3.88NDD24 pKa = 3.64KK25 pKa = 10.97IAMRR29 pKa = 11.84NCIHH33 pKa = 6.67MSSKK37 pKa = 10.12MDD39 pKa = 3.38DD40 pKa = 3.42WLKK43 pKa = 10.18TDD45 pKa = 3.62YY46 pKa = 9.72EE47 pKa = 4.43LCVFPIEE54 pKa = 4.69FYY56 pKa = 11.34DD57 pKa = 4.24DD58 pKa = 3.82FGHH61 pKa = 6.89FAVLHH66 pKa = 6.14DD67 pKa = 4.24LCKK70 pKa = 11.06GNIIPYY76 pKa = 10.15DD77 pKa = 3.73VICNFSEE84 pKa = 5.44LSDD87 pKa = 3.53KK88 pKa = 11.37YY89 pKa = 11.56NLVEE93 pKa = 4.96LNEE96 pKa = 4.12QTEE99 pKa = 4.39DD100 pKa = 3.2
MM1 pKa = 7.42KK2 pKa = 10.33KK3 pKa = 10.42LYY5 pKa = 9.56TIHH8 pKa = 7.64DD9 pKa = 4.63KK10 pKa = 10.77IANKK14 pKa = 10.5CGDD17 pKa = 3.32IFTAEE22 pKa = 3.88NDD24 pKa = 3.64KK25 pKa = 10.97IAMRR29 pKa = 11.84NCIHH33 pKa = 6.67MSSKK37 pKa = 10.12MDD39 pKa = 3.38DD40 pKa = 3.42WLKK43 pKa = 10.18TDD45 pKa = 3.62YY46 pKa = 9.72EE47 pKa = 4.43LCVFPIEE54 pKa = 4.69FYY56 pKa = 11.34DD57 pKa = 4.24DD58 pKa = 3.82FGHH61 pKa = 6.89FAVLHH66 pKa = 6.14DD67 pKa = 4.24LCKK70 pKa = 11.06GNIIPYY76 pKa = 10.15DD77 pKa = 3.73VICNFSEE84 pKa = 5.44LSDD87 pKa = 3.53KK88 pKa = 11.37YY89 pKa = 11.56NLVEE93 pKa = 4.96LNEE96 pKa = 4.12QTEE99 pKa = 4.39DD100 pKa = 3.2
Molecular weight: 11.74 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W5F8|A0A4P8W5F8_9VIRU Major capsid protein OS=Capybara microvirus Cap1_SP_216 OX=2585411 PE=3 SV=1
MM1 pKa = 7.63ALGFNCLDD9 pKa = 3.84CFHH12 pKa = 6.87TRR14 pKa = 11.84TIKK17 pKa = 10.83RR18 pKa = 11.84PDD20 pKa = 3.46NGSPMVVPCGNCMACRR36 pKa = 11.84IARR39 pKa = 11.84SRR41 pKa = 11.84EE42 pKa = 3.32WSLRR46 pKa = 11.84LLMEE50 pKa = 5.05LEE52 pKa = 3.91SHH54 pKa = 6.8DD55 pKa = 4.36FKK57 pKa = 11.71GCFITLTYY65 pKa = 10.92DD66 pKa = 4.76DD67 pKa = 4.14EE68 pKa = 5.02HH69 pKa = 8.22LPNDD73 pKa = 3.81YY74 pKa = 10.93GLHH77 pKa = 6.63KK78 pKa = 10.65EE79 pKa = 4.08DD80 pKa = 5.77LKK82 pKa = 11.56NFWKK86 pKa = 10.68RR87 pKa = 11.84LRR89 pKa = 11.84KK90 pKa = 10.1DD91 pKa = 2.96LDD93 pKa = 3.75FKK95 pKa = 10.87IKK97 pKa = 10.58YY98 pKa = 6.86YY99 pKa = 11.17ACGEE103 pKa = 4.11YY104 pKa = 10.53GDD106 pKa = 4.28TYY108 pKa = 11.23GRR110 pKa = 11.84PHH112 pKa = 5.37YY113 pKa = 10.04HH114 pKa = 7.1AIIFGLDD121 pKa = 3.11PDD123 pKa = 4.34DD124 pKa = 3.44FTRR127 pKa = 11.84RR128 pKa = 11.84LLVDD132 pKa = 3.14NWRR135 pKa = 11.84FCSPDD140 pKa = 2.99RR141 pKa = 11.84FNGTKK146 pKa = 9.88QGLADD151 pKa = 3.69VTIDD155 pKa = 3.44SINYY159 pKa = 4.03VTGYY163 pKa = 9.15IQKK166 pKa = 10.33KK167 pKa = 9.43YY168 pKa = 10.91NGDD171 pKa = 3.69LAKK174 pKa = 10.21EE175 pKa = 4.58VYY177 pKa = 10.67GEE179 pKa = 4.01LQPPFSSSSQGLGLEE194 pKa = 4.24TFLKK198 pKa = 10.22NRR200 pKa = 11.84EE201 pKa = 3.86RR202 pKa = 11.84FEE204 pKa = 4.18NDD206 pKa = 3.11GYY208 pKa = 10.65IGYY211 pKa = 9.77QGVKK215 pKa = 7.76YY216 pKa = 8.3TIPRR220 pKa = 11.84YY221 pKa = 9.2FIKK224 pKa = 10.69KK225 pKa = 9.94LDD227 pKa = 3.81LEE229 pKa = 4.43VSKK232 pKa = 11.08VYY234 pKa = 9.6STEE237 pKa = 3.75KK238 pKa = 9.56VNEE241 pKa = 3.95YY242 pKa = 10.62LEE244 pKa = 4.57KK245 pKa = 11.15NGIDD249 pKa = 3.23PSRR252 pKa = 11.84FVKK255 pKa = 9.61ATSPNFIKK263 pKa = 10.66NVAYY267 pKa = 10.58HH268 pKa = 6.31EE269 pKa = 4.41VADD272 pKa = 3.8PHH274 pKa = 6.67LAIAEE279 pKa = 4.05RR280 pKa = 11.84RR281 pKa = 11.84LLRR284 pKa = 11.84QKK286 pKa = 10.66QLFEE290 pKa = 4.29KK291 pKa = 10.65DD292 pKa = 3.0KK293 pKa = 11.56LKK295 pKa = 11.15
MM1 pKa = 7.63ALGFNCLDD9 pKa = 3.84CFHH12 pKa = 6.87TRR14 pKa = 11.84TIKK17 pKa = 10.83RR18 pKa = 11.84PDD20 pKa = 3.46NGSPMVVPCGNCMACRR36 pKa = 11.84IARR39 pKa = 11.84SRR41 pKa = 11.84EE42 pKa = 3.32WSLRR46 pKa = 11.84LLMEE50 pKa = 5.05LEE52 pKa = 3.91SHH54 pKa = 6.8DD55 pKa = 4.36FKK57 pKa = 11.71GCFITLTYY65 pKa = 10.92DD66 pKa = 4.76DD67 pKa = 4.14EE68 pKa = 5.02HH69 pKa = 8.22LPNDD73 pKa = 3.81YY74 pKa = 10.93GLHH77 pKa = 6.63KK78 pKa = 10.65EE79 pKa = 4.08DD80 pKa = 5.77LKK82 pKa = 11.56NFWKK86 pKa = 10.68RR87 pKa = 11.84LRR89 pKa = 11.84KK90 pKa = 10.1DD91 pKa = 2.96LDD93 pKa = 3.75FKK95 pKa = 10.87IKK97 pKa = 10.58YY98 pKa = 6.86YY99 pKa = 11.17ACGEE103 pKa = 4.11YY104 pKa = 10.53GDD106 pKa = 4.28TYY108 pKa = 11.23GRR110 pKa = 11.84PHH112 pKa = 5.37YY113 pKa = 10.04HH114 pKa = 7.1AIIFGLDD121 pKa = 3.11PDD123 pKa = 4.34DD124 pKa = 3.44FTRR127 pKa = 11.84RR128 pKa = 11.84LLVDD132 pKa = 3.14NWRR135 pKa = 11.84FCSPDD140 pKa = 2.99RR141 pKa = 11.84FNGTKK146 pKa = 9.88QGLADD151 pKa = 3.69VTIDD155 pKa = 3.44SINYY159 pKa = 4.03VTGYY163 pKa = 9.15IQKK166 pKa = 10.33KK167 pKa = 9.43YY168 pKa = 10.91NGDD171 pKa = 3.69LAKK174 pKa = 10.21EE175 pKa = 4.58VYY177 pKa = 10.67GEE179 pKa = 4.01LQPPFSSSSQGLGLEE194 pKa = 4.24TFLKK198 pKa = 10.22NRR200 pKa = 11.84EE201 pKa = 3.86RR202 pKa = 11.84FEE204 pKa = 4.18NDD206 pKa = 3.11GYY208 pKa = 10.65IGYY211 pKa = 9.77QGVKK215 pKa = 7.76YY216 pKa = 8.3TIPRR220 pKa = 11.84YY221 pKa = 9.2FIKK224 pKa = 10.69KK225 pKa = 9.94LDD227 pKa = 3.81LEE229 pKa = 4.43VSKK232 pKa = 11.08VYY234 pKa = 9.6STEE237 pKa = 3.75KK238 pKa = 9.56VNEE241 pKa = 3.95YY242 pKa = 10.62LEE244 pKa = 4.57KK245 pKa = 11.15NGIDD249 pKa = 3.23PSRR252 pKa = 11.84FVKK255 pKa = 9.61ATSPNFIKK263 pKa = 10.66NVAYY267 pKa = 10.58HH268 pKa = 6.31EE269 pKa = 4.41VADD272 pKa = 3.8PHH274 pKa = 6.67LAIAEE279 pKa = 4.05RR280 pKa = 11.84RR281 pKa = 11.84LLRR284 pKa = 11.84QKK286 pKa = 10.66QLFEE290 pKa = 4.29KK291 pKa = 10.65DD292 pKa = 3.0KK293 pKa = 11.56LKK295 pKa = 11.15
Molecular weight: 34.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1406 |
74 |
559 |
234.3 |
26.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.117 ± 1.386 | 1.351 ± 0.539 |
6.472 ± 0.97 | 6.615 ± 0.808 |
5.334 ± 0.565 | 6.686 ± 0.732 |
2.134 ± 0.259 | 5.69 ± 0.628 |
6.401 ± 0.905 | 8.606 ± 0.328 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.418 ± 0.429 | 7.183 ± 0.688 |
3.698 ± 0.653 | 3.698 ± 0.775 |
4.41 ± 0.866 | 6.828 ± 0.888 |
4.836 ± 0.448 | 5.548 ± 0.369 |
1.138 ± 0.26 | 4.836 ± 0.415 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
