
Cricetid gammaherpesvirus 2
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Gammaherpesvirinae; Rhadinovirus
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins
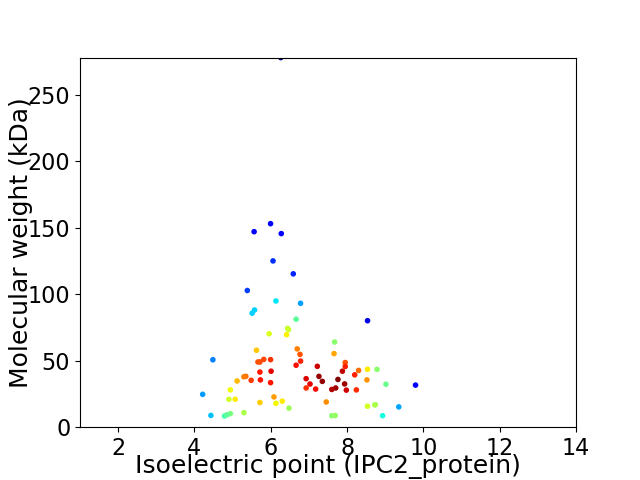
Virtual 2D-PAGE plot for 82 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
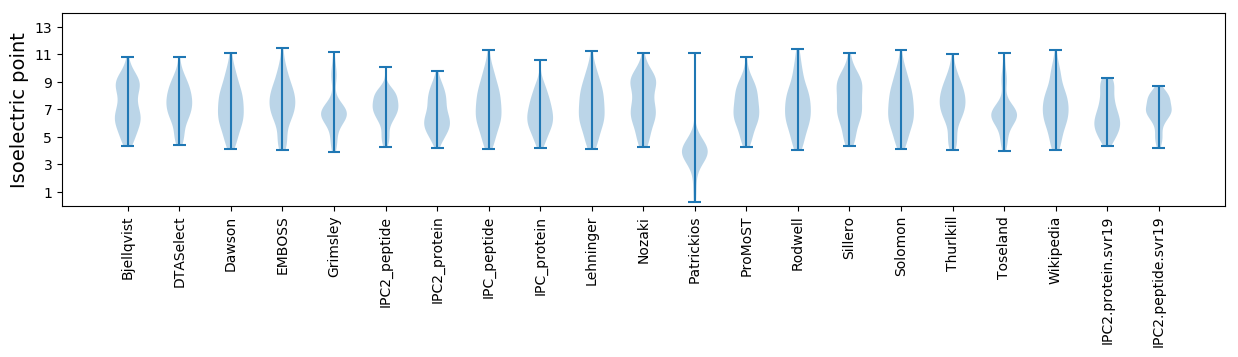
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E9M5L4|E9M5L4_9GAMA Uncharacterized protein OS=Cricetid gammaherpesvirus 2 OX=1605972 GN=RHVP-L.31 PE=3 SV=1
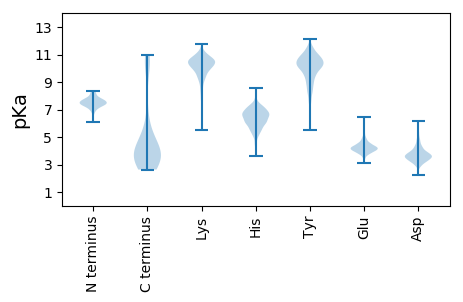
MM1 pKa = 7.59SGPEE5 pKa = 3.72CSEE8 pKa = 4.05KK9 pKa = 10.64KK10 pKa = 10.33KK11 pKa = 9.6EE12 pKa = 4.26TISDD16 pKa = 3.58NDD18 pKa = 3.36MEE20 pKa = 4.4EE21 pKa = 4.19CMKK24 pKa = 10.43FFTMPLPQMLHH35 pKa = 7.33AINQSIADD43 pKa = 3.76FEE45 pKa = 4.84IVCSASQQLEE55 pKa = 4.29RR56 pKa = 11.84ACLLFDD62 pKa = 4.14FVGAEE67 pKa = 4.22CVKK70 pKa = 10.57DD71 pKa = 3.6VGMEE75 pKa = 3.84THH77 pKa = 7.17AEE79 pKa = 3.86
MM1 pKa = 7.59SGPEE5 pKa = 3.72CSEE8 pKa = 4.05KK9 pKa = 10.64KK10 pKa = 10.33KK11 pKa = 9.6EE12 pKa = 4.26TISDD16 pKa = 3.58NDD18 pKa = 3.36MEE20 pKa = 4.4EE21 pKa = 4.19CMKK24 pKa = 10.43FFTMPLPQMLHH35 pKa = 7.33AINQSIADD43 pKa = 3.76FEE45 pKa = 4.84IVCSASQQLEE55 pKa = 4.29RR56 pKa = 11.84ACLLFDD62 pKa = 4.14FVGAEE67 pKa = 4.22CVKK70 pKa = 10.57DD71 pKa = 3.6VGMEE75 pKa = 3.84THH77 pKa = 7.17AEE79 pKa = 3.86
Molecular weight: 8.87 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E9M5K4|E9M5K4_9GAMA Thymidine kinase OS=Cricetid gammaherpesvirus 2 OX=1605972 GN=RHVP-L.21 PE=3 SV=1
MM1 pKa = 7.77AALHH5 pKa = 6.38KK6 pKa = 10.48NWAKK10 pKa = 8.73QTLAQLPPKK19 pKa = 9.88RR20 pKa = 11.84KK21 pKa = 9.28AVGKK25 pKa = 9.56RR26 pKa = 11.84AHH28 pKa = 6.22LRR30 pKa = 11.84CYY32 pKa = 9.84IQLLRR37 pKa = 11.84EE38 pKa = 4.28PTLHH42 pKa = 7.46ALLDD46 pKa = 3.7QLKK49 pKa = 10.0ISHH52 pKa = 7.1TDD54 pKa = 2.88GCHH57 pKa = 5.9QLFKK61 pKa = 11.0LYY63 pKa = 10.71FEE65 pKa = 4.53VRR67 pKa = 11.84LGHH70 pKa = 6.31RR71 pKa = 11.84VADD74 pKa = 4.76AIILTGNDD82 pKa = 4.22SLCHH86 pKa = 6.12CYY88 pKa = 10.0ILEE91 pKa = 4.49FKK93 pKa = 9.79TCASPKK99 pKa = 9.98FDD101 pKa = 3.75INCQIRR107 pKa = 11.84AAQHH111 pKa = 6.88AEE113 pKa = 4.0GCSQLRR119 pKa = 11.84DD120 pKa = 3.86MLSHH124 pKa = 6.76LRR126 pKa = 11.84WHH128 pKa = 6.33VPAGPEE134 pKa = 3.89SCSISCHH141 pKa = 6.43LVFKK145 pKa = 9.38SQRR148 pKa = 11.84GLKK151 pKa = 8.35TLYY154 pKa = 10.44SKK156 pKa = 11.13SLFGASASIPASRR169 pKa = 11.84SALHH173 pKa = 6.12VLLSSLEE180 pKa = 3.79DD181 pKa = 2.98RR182 pKa = 11.84AFKK185 pKa = 9.76NTLLRR190 pKa = 11.84HH191 pKa = 4.88VARR194 pKa = 11.84AKK196 pKa = 10.39HH197 pKa = 5.84DD198 pKa = 3.61RR199 pKa = 11.84CNLLGPKK206 pKa = 8.1SQKK209 pKa = 8.53HH210 pKa = 3.77TAPRR214 pKa = 11.84PKK216 pKa = 9.77HH217 pKa = 5.0PKK219 pKa = 8.67PRR221 pKa = 11.84KK222 pKa = 8.0CAGRR226 pKa = 11.84SALGQTGLRR235 pKa = 11.84TPNPRR240 pKa = 11.84KK241 pKa = 9.78KK242 pKa = 10.24QPPKK246 pKa = 9.98NRR248 pKa = 11.84ARR250 pKa = 11.84PRR252 pKa = 11.84RR253 pKa = 11.84AFPADD258 pKa = 3.1TKK260 pKa = 10.52QRR262 pKa = 11.84SRR264 pKa = 11.84ARR266 pKa = 11.84AGSSQRR272 pKa = 11.84IPKK275 pKa = 8.64GHH277 pKa = 7.57KK278 pKa = 9.21SRR280 pKa = 11.84KK281 pKa = 7.71SS282 pKa = 3.17
MM1 pKa = 7.77AALHH5 pKa = 6.38KK6 pKa = 10.48NWAKK10 pKa = 8.73QTLAQLPPKK19 pKa = 9.88RR20 pKa = 11.84KK21 pKa = 9.28AVGKK25 pKa = 9.56RR26 pKa = 11.84AHH28 pKa = 6.22LRR30 pKa = 11.84CYY32 pKa = 9.84IQLLRR37 pKa = 11.84EE38 pKa = 4.28PTLHH42 pKa = 7.46ALLDD46 pKa = 3.7QLKK49 pKa = 10.0ISHH52 pKa = 7.1TDD54 pKa = 2.88GCHH57 pKa = 5.9QLFKK61 pKa = 11.0LYY63 pKa = 10.71FEE65 pKa = 4.53VRR67 pKa = 11.84LGHH70 pKa = 6.31RR71 pKa = 11.84VADD74 pKa = 4.76AIILTGNDD82 pKa = 4.22SLCHH86 pKa = 6.12CYY88 pKa = 10.0ILEE91 pKa = 4.49FKK93 pKa = 9.79TCASPKK99 pKa = 9.98FDD101 pKa = 3.75INCQIRR107 pKa = 11.84AAQHH111 pKa = 6.88AEE113 pKa = 4.0GCSQLRR119 pKa = 11.84DD120 pKa = 3.86MLSHH124 pKa = 6.76LRR126 pKa = 11.84WHH128 pKa = 6.33VPAGPEE134 pKa = 3.89SCSISCHH141 pKa = 6.43LVFKK145 pKa = 9.38SQRR148 pKa = 11.84GLKK151 pKa = 8.35TLYY154 pKa = 10.44SKK156 pKa = 11.13SLFGASASIPASRR169 pKa = 11.84SALHH173 pKa = 6.12VLLSSLEE180 pKa = 3.79DD181 pKa = 2.98RR182 pKa = 11.84AFKK185 pKa = 9.76NTLLRR190 pKa = 11.84HH191 pKa = 4.88VARR194 pKa = 11.84AKK196 pKa = 10.39HH197 pKa = 5.84DD198 pKa = 3.61RR199 pKa = 11.84CNLLGPKK206 pKa = 8.1SQKK209 pKa = 8.53HH210 pKa = 3.77TAPRR214 pKa = 11.84PKK216 pKa = 9.77HH217 pKa = 5.0PKK219 pKa = 8.67PRR221 pKa = 11.84KK222 pKa = 8.0CAGRR226 pKa = 11.84SALGQTGLRR235 pKa = 11.84TPNPRR240 pKa = 11.84KK241 pKa = 9.78KK242 pKa = 10.24QPPKK246 pKa = 9.98NRR248 pKa = 11.84ARR250 pKa = 11.84PRR252 pKa = 11.84RR253 pKa = 11.84AFPADD258 pKa = 3.1TKK260 pKa = 10.52QRR262 pKa = 11.84SRR264 pKa = 11.84ARR266 pKa = 11.84AGSSQRR272 pKa = 11.84IPKK275 pKa = 8.64GHH277 pKa = 7.57KK278 pKa = 9.21SRR280 pKa = 11.84KK281 pKa = 7.71SS282 pKa = 3.17
Molecular weight: 31.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
35689 |
77 |
2505 |
435.2 |
48.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.094 ± 0.202 | 2.88 ± 0.195 |
4.62 ± 0.101 | 5.114 ± 0.157 |
4.363 ± 0.166 | 5.167 ± 0.251 |
2.88 ± 0.127 | 6.218 ± 0.277 |
5.548 ± 0.214 | 10.39 ± 0.263 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.634 ± 0.131 | 4.36 ± 0.142 |
6.136 ± 0.318 | 4.046 ± 0.142 |
4.122 ± 0.245 | 8.235 ± 0.281 |
6.901 ± 0.276 | 5.898 ± 0.272 |
1.132 ± 0.073 | 3.262 ± 0.119 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
