
Cellulomonas bogoriensis 69B4 = DSM 16987
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Cellulomonadaceae; Cellulomonas; Cellulomonas bogoriensis
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
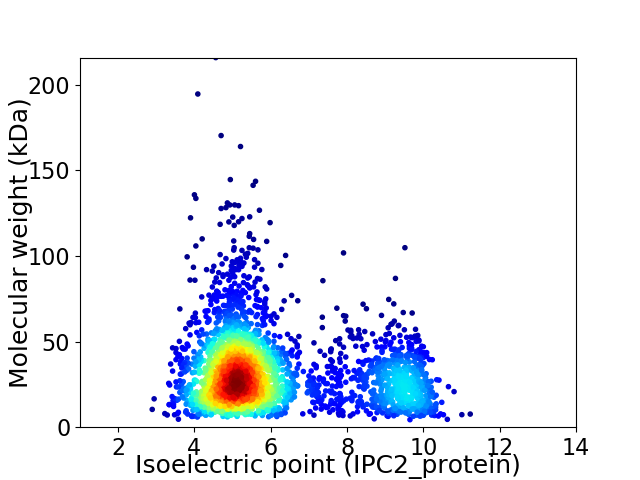
Virtual 2D-PAGE plot for 2898 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A0C0Q4|A0A0A0C0Q4_9CELL Iron ABC transporter permease OS=Cellulomonas bogoriensis 69B4 = DSM 16987 OX=1386082 GN=N869_13145 PE=3 SV=1
MM1 pKa = 7.35RR2 pKa = 11.84TTRR5 pKa = 11.84NRR7 pKa = 11.84GLTAVAAGGFALALTLAACTPPPEE31 pKa = 5.15DD32 pKa = 3.94SPAGPGDD39 pKa = 4.22DD40 pKa = 4.14GDD42 pKa = 4.14RR43 pKa = 11.84DD44 pKa = 3.59LAEE47 pKa = 4.17GCEE50 pKa = 4.0AFEE53 pKa = 4.96QYY55 pKa = 11.37GDD57 pKa = 3.72LSGTSVSVYY66 pKa = 10.12TSITAPEE73 pKa = 4.44DD74 pKa = 3.42QPHH77 pKa = 6.5IDD79 pKa = 3.46SYY81 pKa = 11.37TEE83 pKa = 3.7FEE85 pKa = 4.54RR86 pKa = 11.84CTGADD91 pKa = 2.59IVYY94 pKa = 10.2DD95 pKa = 3.72GSRR98 pKa = 11.84EE99 pKa = 3.9FEE101 pKa = 3.89AQLPVRR107 pKa = 11.84VQAGNPPDD115 pKa = 3.36IAYY118 pKa = 9.43IPQPGLLQTIVRR130 pKa = 11.84DD131 pKa = 4.05FPDD134 pKa = 3.15AVVPVGDD141 pKa = 3.84LAEE144 pKa = 4.66SNVDD148 pKa = 3.42EE149 pKa = 5.2YY150 pKa = 11.9YY151 pKa = 10.83DD152 pKa = 4.3PSWKK156 pKa = 10.32EE157 pKa = 3.59YY158 pKa = 8.79GTVDD162 pKa = 3.04GTYY165 pKa = 10.15YY166 pKa = 9.98ATPLGANVKK175 pKa = 10.15SFVWYY180 pKa = 10.67SPDD183 pKa = 3.03AFADD187 pKa = 3.26AGYY190 pKa = 9.23EE191 pKa = 4.2IPEE194 pKa = 4.18TWDD197 pKa = 3.14EE198 pKa = 4.4LLALSDD204 pKa = 4.58QIVDD208 pKa = 3.94DD209 pKa = 6.05GGTPWCVGIEE219 pKa = 4.16SGDD222 pKa = 3.55ATGWPATDD230 pKa = 3.1WLEE233 pKa = 4.49DD234 pKa = 3.53VMLRR238 pKa = 11.84TAGPEE243 pKa = 4.35VYY245 pKa = 10.05DD246 pKa = 3.02QWVDD250 pKa = 3.35HH251 pKa = 6.95EE252 pKa = 5.18IPFNDD257 pKa = 3.82PAVAEE262 pKa = 4.07ALAAVGEE269 pKa = 4.2ILKK272 pKa = 10.84NEE274 pKa = 4.46DD275 pKa = 3.22YY276 pKa = 11.63VNAGFGDD283 pKa = 3.74VRR285 pKa = 11.84SIASTPFTDD294 pKa = 3.93AGFPILDD301 pKa = 3.99GEE303 pKa = 4.67CWMHH307 pKa = 6.24RR308 pKa = 11.84QASFYY313 pKa = 9.87AANWEE318 pKa = 4.31SADD321 pKa = 3.58PDD323 pKa = 3.88VQVGEE328 pKa = 4.39NGDD331 pKa = 3.52VFAFYY336 pKa = 10.94LPGATTDD343 pKa = 3.52EE344 pKa = 4.46SPLLGGGEE352 pKa = 3.98FVAAFDD358 pKa = 4.56EE359 pKa = 4.72RR360 pKa = 11.84PEE362 pKa = 4.05VQAFQAFLTSPEE374 pKa = 3.95WVNAKK379 pKa = 9.96IPEE382 pKa = 4.47HH383 pKa = 7.17PDD385 pKa = 3.48GGWVSANRR393 pKa = 11.84GQDD396 pKa = 3.25MEE398 pKa = 5.72LYY400 pKa = 10.42ASPIDD405 pKa = 3.74RR406 pKa = 11.84LSAEE410 pKa = 4.59LLADD414 pKa = 4.28DD415 pKa = 5.12DD416 pKa = 4.42AVFRR420 pKa = 11.84FDD422 pKa = 5.94ASDD425 pKa = 3.65LMPGEE430 pKa = 4.43VGSNSFWTEE439 pKa = 3.5MVNWIANDD447 pKa = 3.85KK448 pKa = 10.21PDD450 pKa = 4.48QDD452 pKa = 3.88VLDD455 pKa = 5.65DD456 pKa = 5.88IEE458 pKa = 5.06ASWPP462 pKa = 3.5
MM1 pKa = 7.35RR2 pKa = 11.84TTRR5 pKa = 11.84NRR7 pKa = 11.84GLTAVAAGGFALALTLAACTPPPEE31 pKa = 5.15DD32 pKa = 3.94SPAGPGDD39 pKa = 4.22DD40 pKa = 4.14GDD42 pKa = 4.14RR43 pKa = 11.84DD44 pKa = 3.59LAEE47 pKa = 4.17GCEE50 pKa = 4.0AFEE53 pKa = 4.96QYY55 pKa = 11.37GDD57 pKa = 3.72LSGTSVSVYY66 pKa = 10.12TSITAPEE73 pKa = 4.44DD74 pKa = 3.42QPHH77 pKa = 6.5IDD79 pKa = 3.46SYY81 pKa = 11.37TEE83 pKa = 3.7FEE85 pKa = 4.54RR86 pKa = 11.84CTGADD91 pKa = 2.59IVYY94 pKa = 10.2DD95 pKa = 3.72GSRR98 pKa = 11.84EE99 pKa = 3.9FEE101 pKa = 3.89AQLPVRR107 pKa = 11.84VQAGNPPDD115 pKa = 3.36IAYY118 pKa = 9.43IPQPGLLQTIVRR130 pKa = 11.84DD131 pKa = 4.05FPDD134 pKa = 3.15AVVPVGDD141 pKa = 3.84LAEE144 pKa = 4.66SNVDD148 pKa = 3.42EE149 pKa = 5.2YY150 pKa = 11.9YY151 pKa = 10.83DD152 pKa = 4.3PSWKK156 pKa = 10.32EE157 pKa = 3.59YY158 pKa = 8.79GTVDD162 pKa = 3.04GTYY165 pKa = 10.15YY166 pKa = 9.98ATPLGANVKK175 pKa = 10.15SFVWYY180 pKa = 10.67SPDD183 pKa = 3.03AFADD187 pKa = 3.26AGYY190 pKa = 9.23EE191 pKa = 4.2IPEE194 pKa = 4.18TWDD197 pKa = 3.14EE198 pKa = 4.4LLALSDD204 pKa = 4.58QIVDD208 pKa = 3.94DD209 pKa = 6.05GGTPWCVGIEE219 pKa = 4.16SGDD222 pKa = 3.55ATGWPATDD230 pKa = 3.1WLEE233 pKa = 4.49DD234 pKa = 3.53VMLRR238 pKa = 11.84TAGPEE243 pKa = 4.35VYY245 pKa = 10.05DD246 pKa = 3.02QWVDD250 pKa = 3.35HH251 pKa = 6.95EE252 pKa = 5.18IPFNDD257 pKa = 3.82PAVAEE262 pKa = 4.07ALAAVGEE269 pKa = 4.2ILKK272 pKa = 10.84NEE274 pKa = 4.46DD275 pKa = 3.22YY276 pKa = 11.63VNAGFGDD283 pKa = 3.74VRR285 pKa = 11.84SIASTPFTDD294 pKa = 3.93AGFPILDD301 pKa = 3.99GEE303 pKa = 4.67CWMHH307 pKa = 6.24RR308 pKa = 11.84QASFYY313 pKa = 9.87AANWEE318 pKa = 4.31SADD321 pKa = 3.58PDD323 pKa = 3.88VQVGEE328 pKa = 4.39NGDD331 pKa = 3.52VFAFYY336 pKa = 10.94LPGATTDD343 pKa = 3.52EE344 pKa = 4.46SPLLGGGEE352 pKa = 3.98FVAAFDD358 pKa = 4.56EE359 pKa = 4.72RR360 pKa = 11.84PEE362 pKa = 4.05VQAFQAFLTSPEE374 pKa = 3.95WVNAKK379 pKa = 9.96IPEE382 pKa = 4.47HH383 pKa = 7.17PDD385 pKa = 3.48GGWVSANRR393 pKa = 11.84GQDD396 pKa = 3.25MEE398 pKa = 5.72LYY400 pKa = 10.42ASPIDD405 pKa = 3.74RR406 pKa = 11.84LSAEE410 pKa = 4.59LLADD414 pKa = 4.28DD415 pKa = 5.12DD416 pKa = 4.42AVFRR420 pKa = 11.84FDD422 pKa = 5.94ASDD425 pKa = 3.65LMPGEE430 pKa = 4.43VGSNSFWTEE439 pKa = 3.5MVNWIANDD447 pKa = 3.85KK448 pKa = 10.21PDD450 pKa = 4.48QDD452 pKa = 3.88VLDD455 pKa = 5.65DD456 pKa = 5.88IEE458 pKa = 5.06ASWPP462 pKa = 3.5
Molecular weight: 50.15 kDa
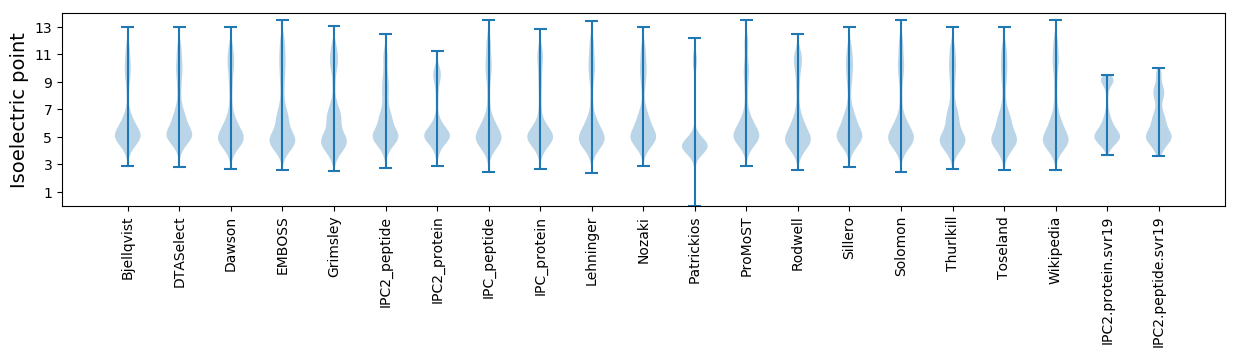
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A0BYF2|A0A0A0BYF2_9CELL Uncharacterized protein OS=Cellulomonas bogoriensis 69B4 = DSM 16987 OX=1386082 GN=N869_14270 PE=4 SV=1
MM1 pKa = 7.7GGPPPPRR8 pKa = 11.84RR9 pKa = 11.84PGRR12 pKa = 11.84PSAGRR17 pKa = 11.84ARR19 pKa = 11.84PGRR22 pKa = 11.84RR23 pKa = 11.84GPGPPRR29 pKa = 11.84PPARR33 pKa = 11.84AAQRR37 pKa = 11.84AARR40 pKa = 11.84VPAAPPRR47 pKa = 11.84APRR50 pKa = 11.84TGPRR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84GRR58 pKa = 11.84RR59 pKa = 11.84RR60 pKa = 11.84ARR62 pKa = 11.84PDD64 pKa = 2.75VTRR67 pKa = 11.84PARR70 pKa = 11.84SGG72 pKa = 3.21
MM1 pKa = 7.7GGPPPPRR8 pKa = 11.84RR9 pKa = 11.84PGRR12 pKa = 11.84PSAGRR17 pKa = 11.84ARR19 pKa = 11.84PGRR22 pKa = 11.84RR23 pKa = 11.84GPGPPRR29 pKa = 11.84PPARR33 pKa = 11.84AAQRR37 pKa = 11.84AARR40 pKa = 11.84VPAAPPRR47 pKa = 11.84APRR50 pKa = 11.84TGPRR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84GRR58 pKa = 11.84RR59 pKa = 11.84RR60 pKa = 11.84ARR62 pKa = 11.84PDD64 pKa = 2.75VTRR67 pKa = 11.84PARR70 pKa = 11.84SGG72 pKa = 3.21
Molecular weight: 7.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
906499 |
37 |
2055 |
312.8 |
33.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.225 ± 0.056 | 0.621 ± 0.012 |
6.423 ± 0.039 | 5.569 ± 0.042 |
2.464 ± 0.027 | 9.529 ± 0.042 |
2.351 ± 0.028 | 2.858 ± 0.033 |
1.299 ± 0.025 | 10.34 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.784 ± 0.02 | 1.504 ± 0.021 |
5.996 ± 0.035 | 2.929 ± 0.023 |
8.028 ± 0.052 | 4.749 ± 0.033 |
6.554 ± 0.038 | 10.49 ± 0.051 |
1.51 ± 0.019 | 1.776 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
