
Demequina mangrovi
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Demequinaceae; Demequina
Average proteome isoelectric point is 5.78
Get precalculated fractions of proteins
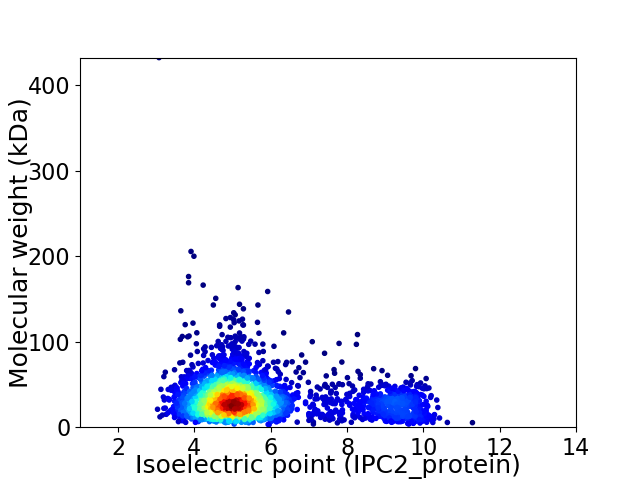
Virtual 2D-PAGE plot for 2745 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H6XH86|A0A1H6XH86_9MICO Ribonucleoside-diphosphate reductase OS=Demequina mangrovi OX=1043493 GN=SAMN05421637_1293 PE=3 SV=1
MM1 pKa = 6.82QRR3 pKa = 11.84RR4 pKa = 11.84TIVPPLVLALAATAVVSAPASAAAPDD30 pKa = 4.15LFISEE35 pKa = 4.53YY36 pKa = 11.08VEE38 pKa = 4.04GSGFNKK44 pKa = 10.02AVEE47 pKa = 4.31IFNGTGAAVDD57 pKa = 3.88LADD60 pKa = 3.86YY61 pKa = 11.14AVVPYY66 pKa = 10.47HH67 pKa = 6.6NGSTDD72 pKa = 3.28PYY74 pKa = 8.47TTIEE78 pKa = 3.95LSGTLEE84 pKa = 4.34PGDD87 pKa = 3.73VFVVASSTTSSTNDD101 pKa = 2.81VAALADD107 pKa = 3.93LTTGSVAWNGNDD119 pKa = 4.09AVLLTHH125 pKa = 7.46DD126 pKa = 4.28GATIDD131 pKa = 3.49VLGTVGDD138 pKa = 4.02DD139 pKa = 4.19SNWGANVTLRR149 pKa = 11.84RR150 pKa = 11.84LEE152 pKa = 4.43SICAGAAAYY161 pKa = 10.11SAGDD165 pKa = 3.42WDD167 pKa = 4.7SYY169 pKa = 11.79ASNTVDD175 pKa = 3.91GLGTHH180 pKa = 6.56TASCDD185 pKa = 3.19GGPVDD190 pKa = 5.56DD191 pKa = 5.49AAPALASSSPEE202 pKa = 4.02DD203 pKa = 3.43GATGVNAAAEE213 pKa = 4.21LTVTFTEE220 pKa = 4.49PVAAEE225 pKa = 3.94GAFALVCDD233 pKa = 4.81DD234 pKa = 3.84ATVPAAVTGGPTTWTVAPEE253 pKa = 4.03TEE255 pKa = 4.09LAAGAVCVLTVTAAAVTDD273 pKa = 4.47LDD275 pKa = 4.01EE276 pKa = 4.76TDD278 pKa = 4.53PPDD281 pKa = 4.43AMEE284 pKa = 4.92ADD286 pKa = 3.52AVIGFTVADD295 pKa = 4.82GPTLIHH301 pKa = 7.11DD302 pKa = 4.01VQGTEE307 pKa = 3.88DD308 pKa = 3.26TAAMVGEE315 pKa = 4.4TVTIEE320 pKa = 4.04GVVVGDD326 pKa = 3.95YY327 pKa = 9.85EE328 pKa = 4.94GPSPTLRR335 pKa = 11.84GFYY338 pKa = 10.03VQEE341 pKa = 3.9EE342 pKa = 4.34DD343 pKa = 4.32ADD345 pKa = 4.08ADD347 pKa = 4.04ADD349 pKa = 4.07PSTSEE354 pKa = 4.83AIFVYY359 pKa = 10.02HH360 pKa = 6.8GSSDD364 pKa = 3.57TVEE367 pKa = 4.29VGDD370 pKa = 4.16LVSVTGTVAEE380 pKa = 4.6RR381 pKa = 11.84FGQTQLGYY389 pKa = 9.85PSALTVLDD397 pKa = 3.63SGLTVTPAAVSLPFADD413 pKa = 3.48ADD415 pKa = 3.54EE416 pKa = 4.67AEE418 pKa = 4.81RR419 pKa = 11.84YY420 pKa = 10.02EE421 pKa = 4.18GMLVEE426 pKa = 5.01FAQTLTVTEE435 pKa = 4.76LYY437 pKa = 10.42EE438 pKa = 3.92LGRR441 pKa = 11.84FGEE444 pKa = 4.65VTVSSGGRR452 pKa = 11.84LDD454 pKa = 3.53QPTAVVEE461 pKa = 4.48PGADD465 pKa = 3.43AIALQEE471 pKa = 4.21ANNLRR476 pKa = 11.84TVIVDD481 pKa = 3.81DD482 pKa = 4.01ATNAQNPDD490 pKa = 3.09VLMGRR495 pKa = 11.84GGDD498 pKa = 3.67PLSAEE503 pKa = 3.86NTLRR507 pKa = 11.84GGDD510 pKa = 4.05TITGLTGVMTFGYY523 pKa = 10.17DD524 pKa = 3.26GYY526 pKa = 11.07YY527 pKa = 10.25SSSNAWRR534 pKa = 11.84VRR536 pKa = 11.84PVDD539 pKa = 3.57ALPAFEE545 pKa = 5.13AVNARR550 pKa = 11.84PTEE553 pKa = 4.06VPDD556 pKa = 3.22VGGDD560 pKa = 3.5VQVASFNVLNYY571 pKa = 10.19FLTLDD576 pKa = 4.69AGDD579 pKa = 4.09NQCGPVGSEE588 pKa = 3.77NDD590 pKa = 4.57CRR592 pKa = 11.84GAWNATEE599 pKa = 5.8LEE601 pKa = 4.16RR602 pKa = 11.84QTDD605 pKa = 3.69KK606 pKa = 11.35LVAALAGLDD615 pKa = 3.47ADD617 pKa = 4.46VIGIMEE623 pKa = 4.45MEE625 pKa = 4.32NTTGVEE631 pKa = 3.77PLAYY635 pKa = 10.21LAEE638 pKa = 4.04QLNAYY643 pKa = 9.89LGTDD647 pKa = 2.73TWSYY651 pKa = 11.26VDD653 pKa = 3.75TGTIGTDD660 pKa = 3.51VIRR663 pKa = 11.84VGILYY668 pKa = 10.39DD669 pKa = 3.49SAVVSEE675 pKa = 4.84LGDD678 pKa = 3.65FAVLDD683 pKa = 3.9STVDD687 pKa = 3.15ARR689 pKa = 11.84FDD691 pKa = 3.46DD692 pKa = 3.91TRR694 pKa = 11.84NRR696 pKa = 11.84PSLAQTFVTEE706 pKa = 4.23DD707 pKa = 3.33DD708 pKa = 3.74AAFTVVTNHH717 pKa = 6.36LKK719 pKa = 10.81SKK721 pKa = 10.24GCSEE725 pKa = 4.52SSGADD730 pKa = 3.16ADD732 pKa = 3.86QGDD735 pKa = 4.95GAGCWNEE742 pKa = 3.78SRR744 pKa = 11.84TLAAEE749 pKa = 4.25ALVDD753 pKa = 3.73WLASDD758 pKa = 3.79PTDD761 pKa = 3.9AHH763 pKa = 6.92DD764 pKa = 5.32ADD766 pKa = 4.24ALVIGDD772 pKa = 3.96LNSYY776 pKa = 10.21GMEE779 pKa = 4.24DD780 pKa = 4.06PIDD783 pKa = 3.7VLRR786 pKa = 11.84EE787 pKa = 3.75AGYY790 pKa = 10.94VEE792 pKa = 5.47LGGGDD797 pKa = 3.63YY798 pKa = 10.97SYY800 pKa = 11.59VFSGQWGSLDD810 pKa = 3.45YY811 pKa = 11.07TFASPSLAAQVTGSAHH827 pKa = 3.8VHH829 pKa = 5.76INADD833 pKa = 3.54EE834 pKa = 4.09PSILDD839 pKa = 3.11YY840 pKa = 7.11TTRR843 pKa = 11.84YY844 pKa = 9.23KK845 pKa = 10.71PSALVEE851 pKa = 4.17SLYY854 pKa = 11.25APDD857 pKa = 3.52WYY859 pKa = 9.59RR860 pKa = 11.84TSDD863 pKa = 3.54HH864 pKa = 7.16DD865 pKa = 4.35PVVLGLDD872 pKa = 3.7LDD874 pKa = 4.31TPVCEE879 pKa = 3.93VSYY882 pKa = 10.71AVNGTWPGGFISQLWIRR899 pKa = 11.84NVSDD903 pKa = 3.65EE904 pKa = 5.54PIDD907 pKa = 3.7GWQLGWDD914 pKa = 3.66FAGDD918 pKa = 4.01EE919 pKa = 4.66EE920 pKa = 5.7VSSLWGGTVAQDD932 pKa = 3.57GAAVTVDD939 pKa = 3.36AMPWNSVLAPVSEE952 pKa = 4.38VKK954 pKa = 10.39GKK956 pKa = 10.39KK957 pKa = 9.57KK958 pKa = 10.89SLGTTLGFLGTSQAGPEE975 pKa = 4.17DD976 pKa = 3.68VTAFTLNGEE985 pKa = 4.3RR986 pKa = 11.84CSVGG990 pKa = 3.09
MM1 pKa = 6.82QRR3 pKa = 11.84RR4 pKa = 11.84TIVPPLVLALAATAVVSAPASAAAPDD30 pKa = 4.15LFISEE35 pKa = 4.53YY36 pKa = 11.08VEE38 pKa = 4.04GSGFNKK44 pKa = 10.02AVEE47 pKa = 4.31IFNGTGAAVDD57 pKa = 3.88LADD60 pKa = 3.86YY61 pKa = 11.14AVVPYY66 pKa = 10.47HH67 pKa = 6.6NGSTDD72 pKa = 3.28PYY74 pKa = 8.47TTIEE78 pKa = 3.95LSGTLEE84 pKa = 4.34PGDD87 pKa = 3.73VFVVASSTTSSTNDD101 pKa = 2.81VAALADD107 pKa = 3.93LTTGSVAWNGNDD119 pKa = 4.09AVLLTHH125 pKa = 7.46DD126 pKa = 4.28GATIDD131 pKa = 3.49VLGTVGDD138 pKa = 4.02DD139 pKa = 4.19SNWGANVTLRR149 pKa = 11.84RR150 pKa = 11.84LEE152 pKa = 4.43SICAGAAAYY161 pKa = 10.11SAGDD165 pKa = 3.42WDD167 pKa = 4.7SYY169 pKa = 11.79ASNTVDD175 pKa = 3.91GLGTHH180 pKa = 6.56TASCDD185 pKa = 3.19GGPVDD190 pKa = 5.56DD191 pKa = 5.49AAPALASSSPEE202 pKa = 4.02DD203 pKa = 3.43GATGVNAAAEE213 pKa = 4.21LTVTFTEE220 pKa = 4.49PVAAEE225 pKa = 3.94GAFALVCDD233 pKa = 4.81DD234 pKa = 3.84ATVPAAVTGGPTTWTVAPEE253 pKa = 4.03TEE255 pKa = 4.09LAAGAVCVLTVTAAAVTDD273 pKa = 4.47LDD275 pKa = 4.01EE276 pKa = 4.76TDD278 pKa = 4.53PPDD281 pKa = 4.43AMEE284 pKa = 4.92ADD286 pKa = 3.52AVIGFTVADD295 pKa = 4.82GPTLIHH301 pKa = 7.11DD302 pKa = 4.01VQGTEE307 pKa = 3.88DD308 pKa = 3.26TAAMVGEE315 pKa = 4.4TVTIEE320 pKa = 4.04GVVVGDD326 pKa = 3.95YY327 pKa = 9.85EE328 pKa = 4.94GPSPTLRR335 pKa = 11.84GFYY338 pKa = 10.03VQEE341 pKa = 3.9EE342 pKa = 4.34DD343 pKa = 4.32ADD345 pKa = 4.08ADD347 pKa = 4.04ADD349 pKa = 4.07PSTSEE354 pKa = 4.83AIFVYY359 pKa = 10.02HH360 pKa = 6.8GSSDD364 pKa = 3.57TVEE367 pKa = 4.29VGDD370 pKa = 4.16LVSVTGTVAEE380 pKa = 4.6RR381 pKa = 11.84FGQTQLGYY389 pKa = 9.85PSALTVLDD397 pKa = 3.63SGLTVTPAAVSLPFADD413 pKa = 3.48ADD415 pKa = 3.54EE416 pKa = 4.67AEE418 pKa = 4.81RR419 pKa = 11.84YY420 pKa = 10.02EE421 pKa = 4.18GMLVEE426 pKa = 5.01FAQTLTVTEE435 pKa = 4.76LYY437 pKa = 10.42EE438 pKa = 3.92LGRR441 pKa = 11.84FGEE444 pKa = 4.65VTVSSGGRR452 pKa = 11.84LDD454 pKa = 3.53QPTAVVEE461 pKa = 4.48PGADD465 pKa = 3.43AIALQEE471 pKa = 4.21ANNLRR476 pKa = 11.84TVIVDD481 pKa = 3.81DD482 pKa = 4.01ATNAQNPDD490 pKa = 3.09VLMGRR495 pKa = 11.84GGDD498 pKa = 3.67PLSAEE503 pKa = 3.86NTLRR507 pKa = 11.84GGDD510 pKa = 4.05TITGLTGVMTFGYY523 pKa = 10.17DD524 pKa = 3.26GYY526 pKa = 11.07YY527 pKa = 10.25SSSNAWRR534 pKa = 11.84VRR536 pKa = 11.84PVDD539 pKa = 3.57ALPAFEE545 pKa = 5.13AVNARR550 pKa = 11.84PTEE553 pKa = 4.06VPDD556 pKa = 3.22VGGDD560 pKa = 3.5VQVASFNVLNYY571 pKa = 10.19FLTLDD576 pKa = 4.69AGDD579 pKa = 4.09NQCGPVGSEE588 pKa = 3.77NDD590 pKa = 4.57CRR592 pKa = 11.84GAWNATEE599 pKa = 5.8LEE601 pKa = 4.16RR602 pKa = 11.84QTDD605 pKa = 3.69KK606 pKa = 11.35LVAALAGLDD615 pKa = 3.47ADD617 pKa = 4.46VIGIMEE623 pKa = 4.45MEE625 pKa = 4.32NTTGVEE631 pKa = 3.77PLAYY635 pKa = 10.21LAEE638 pKa = 4.04QLNAYY643 pKa = 9.89LGTDD647 pKa = 2.73TWSYY651 pKa = 11.26VDD653 pKa = 3.75TGTIGTDD660 pKa = 3.51VIRR663 pKa = 11.84VGILYY668 pKa = 10.39DD669 pKa = 3.49SAVVSEE675 pKa = 4.84LGDD678 pKa = 3.65FAVLDD683 pKa = 3.9STVDD687 pKa = 3.15ARR689 pKa = 11.84FDD691 pKa = 3.46DD692 pKa = 3.91TRR694 pKa = 11.84NRR696 pKa = 11.84PSLAQTFVTEE706 pKa = 4.23DD707 pKa = 3.33DD708 pKa = 3.74AAFTVVTNHH717 pKa = 6.36LKK719 pKa = 10.81SKK721 pKa = 10.24GCSEE725 pKa = 4.52SSGADD730 pKa = 3.16ADD732 pKa = 3.86QGDD735 pKa = 4.95GAGCWNEE742 pKa = 3.78SRR744 pKa = 11.84TLAAEE749 pKa = 4.25ALVDD753 pKa = 3.73WLASDD758 pKa = 3.79PTDD761 pKa = 3.9AHH763 pKa = 6.92DD764 pKa = 5.32ADD766 pKa = 4.24ALVIGDD772 pKa = 3.96LNSYY776 pKa = 10.21GMEE779 pKa = 4.24DD780 pKa = 4.06PIDD783 pKa = 3.7VLRR786 pKa = 11.84EE787 pKa = 3.75AGYY790 pKa = 10.94VEE792 pKa = 5.47LGGGDD797 pKa = 3.63YY798 pKa = 10.97SYY800 pKa = 11.59VFSGQWGSLDD810 pKa = 3.45YY811 pKa = 11.07TFASPSLAAQVTGSAHH827 pKa = 3.8VHH829 pKa = 5.76INADD833 pKa = 3.54EE834 pKa = 4.09PSILDD839 pKa = 3.11YY840 pKa = 7.11TTRR843 pKa = 11.84YY844 pKa = 9.23KK845 pKa = 10.71PSALVEE851 pKa = 4.17SLYY854 pKa = 11.25APDD857 pKa = 3.52WYY859 pKa = 9.59RR860 pKa = 11.84TSDD863 pKa = 3.54HH864 pKa = 7.16DD865 pKa = 4.35PVVLGLDD872 pKa = 3.7LDD874 pKa = 4.31TPVCEE879 pKa = 3.93VSYY882 pKa = 10.71AVNGTWPGGFISQLWIRR899 pKa = 11.84NVSDD903 pKa = 3.65EE904 pKa = 5.54PIDD907 pKa = 3.7GWQLGWDD914 pKa = 3.66FAGDD918 pKa = 4.01EE919 pKa = 4.66EE920 pKa = 5.7VSSLWGGTVAQDD932 pKa = 3.57GAAVTVDD939 pKa = 3.36AMPWNSVLAPVSEE952 pKa = 4.38VKK954 pKa = 10.39GKK956 pKa = 10.39KK957 pKa = 9.57KK958 pKa = 10.89SLGTTLGFLGTSQAGPEE975 pKa = 4.17DD976 pKa = 3.68VTAFTLNGEE985 pKa = 4.3RR986 pKa = 11.84CSVGG990 pKa = 3.09
Molecular weight: 102.8 kDa
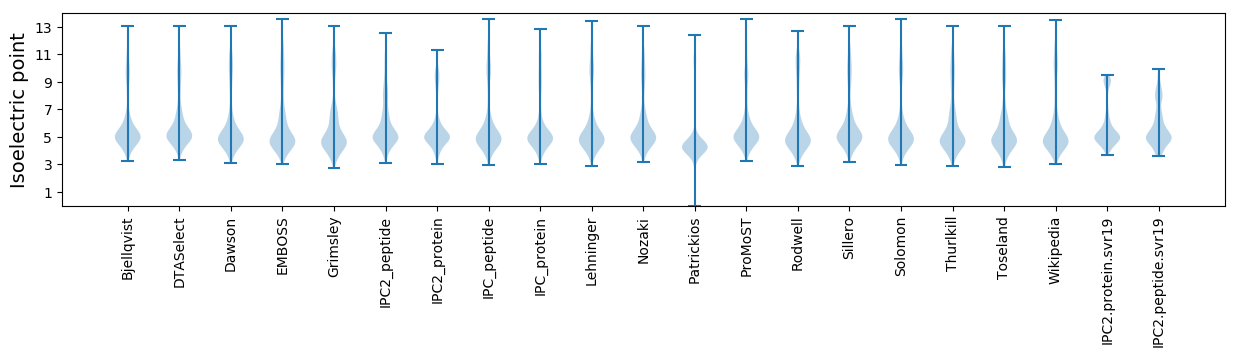
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H6ZWS1|A0A1H6ZWS1_9MICO Chemotaxis protein MotA OS=Demequina mangrovi OX=1043493 GN=SAMN05421637_2194 PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84KK12 pKa = 8.98RR13 pKa = 11.84AKK15 pKa = 8.46THH17 pKa = 5.11GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84SIMASRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 10.21GRR40 pKa = 11.84GKK42 pKa = 10.78LSAA45 pKa = 4.32
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84KK12 pKa = 8.98RR13 pKa = 11.84AKK15 pKa = 8.46THH17 pKa = 5.11GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84SIMASRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 10.21GRR40 pKa = 11.84GKK42 pKa = 10.78LSAA45 pKa = 4.32
Molecular weight: 5.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
912256 |
29 |
4176 |
332.3 |
35.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.526 ± 0.08 | 0.599 ± 0.013 |
6.385 ± 0.054 | 6.292 ± 0.054 |
2.771 ± 0.03 | 9.097 ± 0.047 |
2.072 ± 0.024 | 4.094 ± 0.036 |
1.824 ± 0.035 | 9.855 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.033 ± 0.021 | 1.646 ± 0.028 |
5.27 ± 0.041 | 2.432 ± 0.023 |
7.174 ± 0.065 | 5.236 ± 0.034 |
5.993 ± 0.056 | 9.224 ± 0.043 |
1.525 ± 0.022 | 1.952 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
