
Circovirus-like genome BBC-A
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
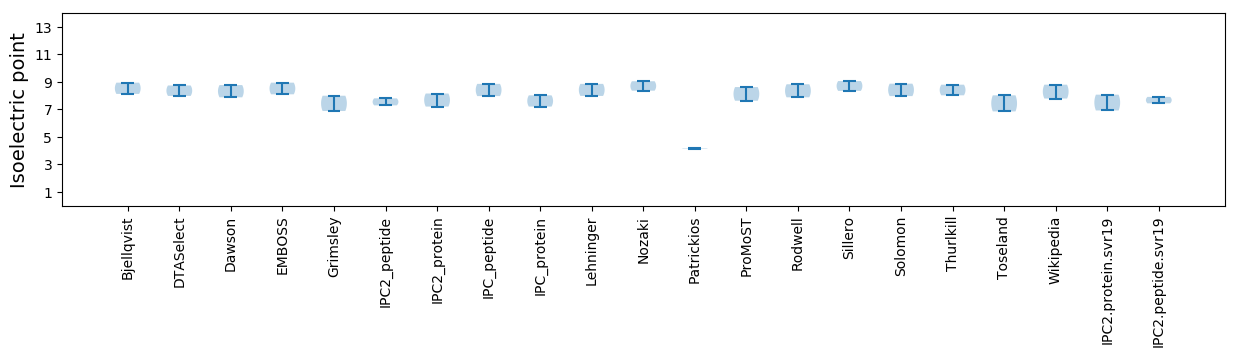
Average proteome isoelectric point is 7.5
Get precalculated fractions of proteins
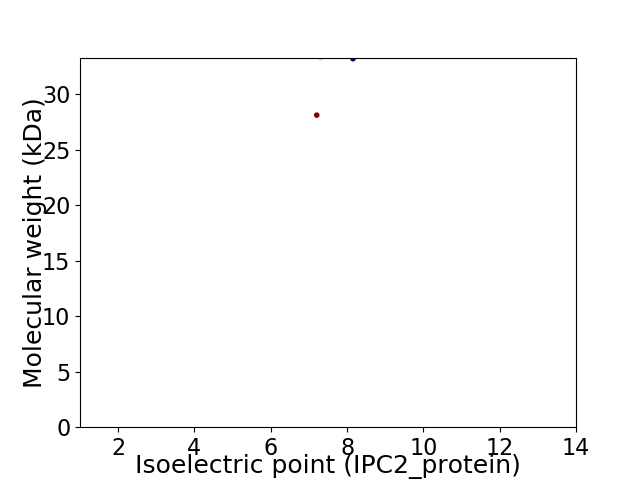
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C6GIJ5|C6GIJ5_9VIRU Uncharacterized protein OS=Circovirus-like genome BBC-A OX=642260 PE=4 SV=1
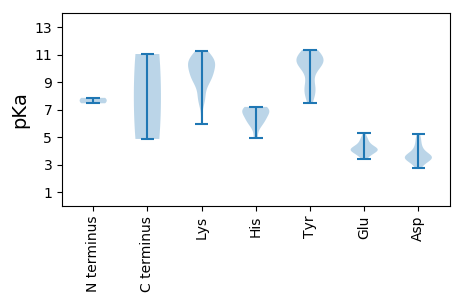
MM1 pKa = 7.46CFSNISHH8 pKa = 7.17INTKK12 pKa = 9.12CHH14 pKa = 7.01HH15 pKa = 6.79EE16 pKa = 4.06NTQKK20 pKa = 11.26KK21 pKa = 9.34NFTKK25 pKa = 10.43KK26 pKa = 8.22VQNVVRR32 pKa = 11.84RR33 pKa = 11.84MAEE36 pKa = 3.99TKK38 pKa = 10.23IQDD41 pKa = 3.32YY42 pKa = 10.77SINEE46 pKa = 4.15DD47 pKa = 3.56VLISNFDD54 pKa = 3.36TPYY57 pKa = 10.87VQDD60 pKa = 3.53LTQIEE65 pKa = 4.31RR66 pKa = 11.84GGASNQRR73 pKa = 11.84IGNEE77 pKa = 3.4ILMSGVKK84 pKa = 8.72WRR86 pKa = 11.84LLFHH90 pKa = 6.87CRR92 pKa = 11.84NNPNYY97 pKa = 9.21EE98 pKa = 4.01DD99 pKa = 3.64VVIRR103 pKa = 11.84SLWCRR108 pKa = 11.84IAIIEE113 pKa = 4.23MKK115 pKa = 9.67NTGTTDD121 pKa = 3.2VVLLEE126 pKa = 4.12DD127 pKa = 4.78FFRR130 pKa = 11.84KK131 pKa = 9.24GANSMDD137 pKa = 3.45FDD139 pKa = 4.19AVTEE143 pKa = 4.08AEE145 pKa = 4.28RR146 pKa = 11.84YY147 pKa = 8.26YY148 pKa = 11.34LPVDD152 pKa = 4.01LTNKK156 pKa = 7.9KK157 pKa = 7.6TLYY160 pKa = 8.88QTTFKK165 pKa = 10.52IGVKK169 pKa = 10.0NQVDD173 pKa = 3.63TADD176 pKa = 3.68YY177 pKa = 10.61LSNKK181 pKa = 9.01IIKK184 pKa = 9.88NYY186 pKa = 10.04RR187 pKa = 11.84KK188 pKa = 9.53VSRR191 pKa = 11.84SINFEE196 pKa = 3.68SSDD199 pKa = 3.33ISNSASTKK207 pKa = 10.42LYY209 pKa = 8.38FVCWIGNNDD218 pKa = 3.46LDD220 pKa = 5.19ANNATTNNAGACEE233 pKa = 3.84LSGLLSTYY241 pKa = 10.95YY242 pKa = 10.85KK243 pKa = 10.92DD244 pKa = 3.59FF245 pKa = 4.86
MM1 pKa = 7.46CFSNISHH8 pKa = 7.17INTKK12 pKa = 9.12CHH14 pKa = 7.01HH15 pKa = 6.79EE16 pKa = 4.06NTQKK20 pKa = 11.26KK21 pKa = 9.34NFTKK25 pKa = 10.43KK26 pKa = 8.22VQNVVRR32 pKa = 11.84RR33 pKa = 11.84MAEE36 pKa = 3.99TKK38 pKa = 10.23IQDD41 pKa = 3.32YY42 pKa = 10.77SINEE46 pKa = 4.15DD47 pKa = 3.56VLISNFDD54 pKa = 3.36TPYY57 pKa = 10.87VQDD60 pKa = 3.53LTQIEE65 pKa = 4.31RR66 pKa = 11.84GGASNQRR73 pKa = 11.84IGNEE77 pKa = 3.4ILMSGVKK84 pKa = 8.72WRR86 pKa = 11.84LLFHH90 pKa = 6.87CRR92 pKa = 11.84NNPNYY97 pKa = 9.21EE98 pKa = 4.01DD99 pKa = 3.64VVIRR103 pKa = 11.84SLWCRR108 pKa = 11.84IAIIEE113 pKa = 4.23MKK115 pKa = 9.67NTGTTDD121 pKa = 3.2VVLLEE126 pKa = 4.12DD127 pKa = 4.78FFRR130 pKa = 11.84KK131 pKa = 9.24GANSMDD137 pKa = 3.45FDD139 pKa = 4.19AVTEE143 pKa = 4.08AEE145 pKa = 4.28RR146 pKa = 11.84YY147 pKa = 8.26YY148 pKa = 11.34LPVDD152 pKa = 4.01LTNKK156 pKa = 7.9KK157 pKa = 7.6TLYY160 pKa = 8.88QTTFKK165 pKa = 10.52IGVKK169 pKa = 10.0NQVDD173 pKa = 3.63TADD176 pKa = 3.68YY177 pKa = 10.61LSNKK181 pKa = 9.01IIKK184 pKa = 9.88NYY186 pKa = 10.04RR187 pKa = 11.84KK188 pKa = 9.53VSRR191 pKa = 11.84SINFEE196 pKa = 3.68SSDD199 pKa = 3.33ISNSASTKK207 pKa = 10.42LYY209 pKa = 8.38FVCWIGNNDD218 pKa = 3.46LDD220 pKa = 5.19ANNATTNNAGACEE233 pKa = 3.84LSGLLSTYY241 pKa = 10.95YY242 pKa = 10.85KK243 pKa = 10.92DD244 pKa = 3.59FF245 pKa = 4.86
Molecular weight: 28.13 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C6GIJ5|C6GIJ5_9VIRU Uncharacterized protein OS=Circovirus-like genome BBC-A OX=642260 PE=4 SV=1
MM1 pKa = 7.85KK2 pKa = 9.41ITTEE6 pKa = 3.73PTKK9 pKa = 10.77RR10 pKa = 11.84RR11 pKa = 11.84DD12 pKa = 3.02WAFTDD17 pKa = 4.87FNMEE21 pKa = 3.89NQTKK25 pKa = 9.32YY26 pKa = 11.2ASLVEE31 pKa = 4.08QCVCSYY37 pKa = 10.56VCFGIEE43 pKa = 3.99YY44 pKa = 9.91CPKK47 pKa = 9.43TNRR50 pKa = 11.84PHH52 pKa = 5.77LQGYY56 pKa = 7.5LHH58 pKa = 6.97FKK60 pKa = 10.15NPRR63 pKa = 11.84TTRR66 pKa = 11.84EE67 pKa = 3.64GRR69 pKa = 11.84NVFGKK74 pKa = 7.55THH76 pKa = 7.19SEE78 pKa = 4.37FCWHH82 pKa = 6.22SRR84 pKa = 11.84EE85 pKa = 4.98KK86 pKa = 10.88NINYY90 pKa = 8.48CKK92 pKa = 10.49KK93 pKa = 7.82GTKK96 pKa = 9.94VLGEE100 pKa = 4.12TFFEE104 pKa = 4.69FGKK107 pKa = 10.66KK108 pKa = 9.22PNQGYY113 pKa = 7.75RR114 pKa = 11.84TDD116 pKa = 3.69IEE118 pKa = 4.13NVEE121 pKa = 4.59YY122 pKa = 10.33ILKK125 pKa = 5.97TTRR128 pKa = 11.84SIQGIIDD135 pKa = 3.35NHH137 pKa = 6.0FNLYY141 pKa = 8.21IQYY144 pKa = 10.46RR145 pKa = 11.84SRR147 pKa = 11.84FLNYY151 pKa = 10.47LEE153 pKa = 4.79DD154 pKa = 3.96TEE156 pKa = 4.49KK157 pKa = 10.77PRR159 pKa = 11.84AEE161 pKa = 3.93MTKK164 pKa = 10.28GIWYY168 pKa = 8.64YY169 pKa = 11.2GPPGCGKK176 pKa = 8.89STVIKK181 pKa = 10.78NNFFGTEE188 pKa = 4.45VVWLEE193 pKa = 3.54YY194 pKa = 10.33DD195 pKa = 3.45GKK197 pKa = 11.18YY198 pKa = 10.5FSDD201 pKa = 4.27YY202 pKa = 10.87NGHH205 pKa = 4.89TTVIFDD211 pKa = 4.48DD212 pKa = 3.63QDD214 pKa = 2.83ISKK217 pKa = 10.15FSRR220 pKa = 11.84SMILKK225 pKa = 9.83LINHH229 pKa = 6.39TPFKK233 pKa = 10.49LRR235 pKa = 11.84CLGSYY240 pKa = 10.4RR241 pKa = 11.84EE242 pKa = 4.19FNAITVIFIDD252 pKa = 3.52NFHH255 pKa = 6.36YY256 pKa = 10.43SHH258 pKa = 6.98YY259 pKa = 10.25FGEE262 pKa = 4.91DD263 pKa = 2.75EE264 pKa = 5.27AIARR268 pKa = 11.84RR269 pKa = 11.84LKK271 pKa = 10.53AIQFGTEE278 pKa = 3.86VPKK281 pKa = 11.04
MM1 pKa = 7.85KK2 pKa = 9.41ITTEE6 pKa = 3.73PTKK9 pKa = 10.77RR10 pKa = 11.84RR11 pKa = 11.84DD12 pKa = 3.02WAFTDD17 pKa = 4.87FNMEE21 pKa = 3.89NQTKK25 pKa = 9.32YY26 pKa = 11.2ASLVEE31 pKa = 4.08QCVCSYY37 pKa = 10.56VCFGIEE43 pKa = 3.99YY44 pKa = 9.91CPKK47 pKa = 9.43TNRR50 pKa = 11.84PHH52 pKa = 5.77LQGYY56 pKa = 7.5LHH58 pKa = 6.97FKK60 pKa = 10.15NPRR63 pKa = 11.84TTRR66 pKa = 11.84EE67 pKa = 3.64GRR69 pKa = 11.84NVFGKK74 pKa = 7.55THH76 pKa = 7.19SEE78 pKa = 4.37FCWHH82 pKa = 6.22SRR84 pKa = 11.84EE85 pKa = 4.98KK86 pKa = 10.88NINYY90 pKa = 8.48CKK92 pKa = 10.49KK93 pKa = 7.82GTKK96 pKa = 9.94VLGEE100 pKa = 4.12TFFEE104 pKa = 4.69FGKK107 pKa = 10.66KK108 pKa = 9.22PNQGYY113 pKa = 7.75RR114 pKa = 11.84TDD116 pKa = 3.69IEE118 pKa = 4.13NVEE121 pKa = 4.59YY122 pKa = 10.33ILKK125 pKa = 5.97TTRR128 pKa = 11.84SIQGIIDD135 pKa = 3.35NHH137 pKa = 6.0FNLYY141 pKa = 8.21IQYY144 pKa = 10.46RR145 pKa = 11.84SRR147 pKa = 11.84FLNYY151 pKa = 10.47LEE153 pKa = 4.79DD154 pKa = 3.96TEE156 pKa = 4.49KK157 pKa = 10.77PRR159 pKa = 11.84AEE161 pKa = 3.93MTKK164 pKa = 10.28GIWYY168 pKa = 8.64YY169 pKa = 11.2GPPGCGKK176 pKa = 8.89STVIKK181 pKa = 10.78NNFFGTEE188 pKa = 4.45VVWLEE193 pKa = 3.54YY194 pKa = 10.33DD195 pKa = 3.45GKK197 pKa = 11.18YY198 pKa = 10.5FSDD201 pKa = 4.27YY202 pKa = 10.87NGHH205 pKa = 4.89TTVIFDD211 pKa = 4.48DD212 pKa = 3.63QDD214 pKa = 2.83ISKK217 pKa = 10.15FSRR220 pKa = 11.84SMILKK225 pKa = 9.83LINHH229 pKa = 6.39TPFKK233 pKa = 10.49LRR235 pKa = 11.84CLGSYY240 pKa = 10.4RR241 pKa = 11.84EE242 pKa = 4.19FNAITVIFIDD252 pKa = 3.52NFHH255 pKa = 6.36YY256 pKa = 10.43SHH258 pKa = 6.98YY259 pKa = 10.25FGEE262 pKa = 4.91DD263 pKa = 2.75EE264 pKa = 5.27AIARR268 pKa = 11.84RR269 pKa = 11.84LKK271 pKa = 10.53AIQFGTEE278 pKa = 3.86VPKK281 pKa = 11.04
Molecular weight: 33.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
526 |
245 |
281 |
263.0 |
30.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.612 ± 0.865 | 2.662 ± 0.143 |
5.323 ± 0.813 | 6.084 ± 0.798 |
6.274 ± 1.2 | 5.513 ± 0.963 |
2.471 ± 0.564 | 7.224 ± 0.083 |
7.795 ± 0.301 | 5.894 ± 0.703 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.711 ± 0.222 | 8.365 ± 1.512 |
2.471 ± 0.839 | 3.042 ± 0.15 |
5.513 ± 0.414 | 5.894 ± 0.978 |
7.985 ± 0.155 | 5.323 ± 0.813 |
1.331 ± 0.072 | 5.513 ± 0.689 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
