
Marinirhabdus gelatinilytica
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Marinirhabdus
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
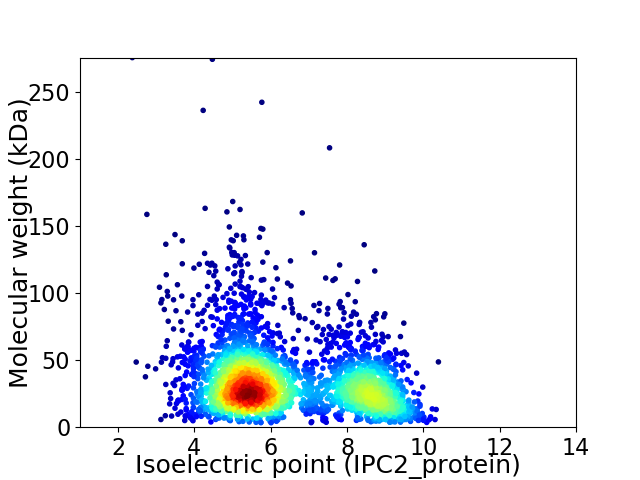
Virtual 2D-PAGE plot for 3022 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A370QGB2|A0A370QGB2_9FLAO Phosphate starvation-inducible protein PhoH OS=Marinirhabdus gelatinilytica OX=1703343 GN=C8D94_102583 PE=4 SV=1
MM1 pKa = 7.48KK2 pKa = 10.4KK3 pKa = 9.77ILLTVLGTLALFSCSKK19 pKa = 10.49DD20 pKa = 3.55SEE22 pKa = 4.66VLTTTSQDD30 pKa = 3.3LTVEE34 pKa = 4.55LAQEE38 pKa = 4.2EE39 pKa = 4.67KK40 pKa = 10.78SVFGTYY46 pKa = 10.17KK47 pKa = 11.02GIFTTNNAEE56 pKa = 3.93YY57 pKa = 10.44RR58 pKa = 11.84GTFEE62 pKa = 4.68LWVPSSNSSSYY73 pKa = 10.76SISAYY78 pKa = 9.81RR79 pKa = 11.84VPTVKK84 pKa = 9.7LTLHH88 pKa = 5.74TGEE91 pKa = 4.89VYY93 pKa = 10.79FSQAEE98 pKa = 4.26SNPVGGRR105 pKa = 11.84ALSNLEE111 pKa = 3.96FVSDD115 pKa = 4.08DD116 pKa = 3.44FTFSFSVNEE125 pKa = 4.62DD126 pKa = 2.94GSNPTVTNVTFKK138 pKa = 10.85NAEE141 pKa = 3.74GDD143 pKa = 3.61MLLAKK148 pKa = 8.89HH149 pKa = 5.37TQRR152 pKa = 11.84APVNPRR158 pKa = 11.84SGTWDD163 pKa = 3.29CTTCNGHH170 pKa = 6.31PVVGQGGTQTFNMMFVTADD189 pKa = 3.45GNSTITTQSTINSTTTDD206 pKa = 4.13GIGVQSNCMANGDD219 pKa = 4.03TTVCTIEE226 pKa = 4.83SGDD229 pKa = 3.7GTTTTTGFLSNGNPVTWLGTHH250 pKa = 6.26TFNNEE255 pKa = 2.97VSGPDD260 pKa = 3.67DD261 pKa = 4.34CSTLQGTWEE270 pKa = 4.28WEE272 pKa = 4.27SISYY276 pKa = 7.71GTLEE280 pKa = 4.45GNFITDD286 pKa = 2.99IAGNCPITDD295 pKa = 3.22VTLINEE301 pKa = 4.44DD302 pKa = 3.94FEE304 pKa = 5.05DD305 pKa = 3.7ATVSYY310 pKa = 10.57VASFDD315 pKa = 4.1DD316 pKa = 5.37LSGEE320 pKa = 3.73NDD322 pKa = 3.52DD323 pKa = 4.92YY324 pKa = 11.74FRR326 pKa = 11.84ITNGTDD332 pKa = 2.73ISATINNVQGSQFFAAQDD350 pKa = 3.56TDD352 pKa = 3.78GVAGNGQSPANLTWSNLNITGLTNINFSAFFAEE385 pKa = 5.45DD386 pKa = 3.79DD387 pKa = 4.12ASDD390 pKa = 4.94AAEE393 pKa = 4.53DD394 pKa = 3.46WDD396 pKa = 3.85NADD399 pKa = 4.36FVDD402 pKa = 3.83VQYY405 pKa = 11.6SFDD408 pKa = 3.43GTTYY412 pKa = 8.02TTFFSIRR419 pKa = 11.84NDD421 pKa = 2.81GSTFNAAPQIDD432 pKa = 4.1TDD434 pKa = 4.32LDD436 pKa = 4.12GVGDD440 pKa = 3.9GAIITDD446 pKa = 3.3NMANYY451 pKa = 9.79SASFPINGTTNPTASTEE468 pKa = 3.47LWIRR472 pKa = 11.84VNVGLDD478 pKa = 3.27SGDD481 pKa = 3.46EE482 pKa = 4.61DD483 pKa = 4.13IAFDD487 pKa = 4.07DD488 pKa = 5.62FIVIGNN494 pKa = 3.95
MM1 pKa = 7.48KK2 pKa = 10.4KK3 pKa = 9.77ILLTVLGTLALFSCSKK19 pKa = 10.49DD20 pKa = 3.55SEE22 pKa = 4.66VLTTTSQDD30 pKa = 3.3LTVEE34 pKa = 4.55LAQEE38 pKa = 4.2EE39 pKa = 4.67KK40 pKa = 10.78SVFGTYY46 pKa = 10.17KK47 pKa = 11.02GIFTTNNAEE56 pKa = 3.93YY57 pKa = 10.44RR58 pKa = 11.84GTFEE62 pKa = 4.68LWVPSSNSSSYY73 pKa = 10.76SISAYY78 pKa = 9.81RR79 pKa = 11.84VPTVKK84 pKa = 9.7LTLHH88 pKa = 5.74TGEE91 pKa = 4.89VYY93 pKa = 10.79FSQAEE98 pKa = 4.26SNPVGGRR105 pKa = 11.84ALSNLEE111 pKa = 3.96FVSDD115 pKa = 4.08DD116 pKa = 3.44FTFSFSVNEE125 pKa = 4.62DD126 pKa = 2.94GSNPTVTNVTFKK138 pKa = 10.85NAEE141 pKa = 3.74GDD143 pKa = 3.61MLLAKK148 pKa = 8.89HH149 pKa = 5.37TQRR152 pKa = 11.84APVNPRR158 pKa = 11.84SGTWDD163 pKa = 3.29CTTCNGHH170 pKa = 6.31PVVGQGGTQTFNMMFVTADD189 pKa = 3.45GNSTITTQSTINSTTTDD206 pKa = 4.13GIGVQSNCMANGDD219 pKa = 4.03TTVCTIEE226 pKa = 4.83SGDD229 pKa = 3.7GTTTTTGFLSNGNPVTWLGTHH250 pKa = 6.26TFNNEE255 pKa = 2.97VSGPDD260 pKa = 3.67DD261 pKa = 4.34CSTLQGTWEE270 pKa = 4.28WEE272 pKa = 4.27SISYY276 pKa = 7.71GTLEE280 pKa = 4.45GNFITDD286 pKa = 2.99IAGNCPITDD295 pKa = 3.22VTLINEE301 pKa = 4.44DD302 pKa = 3.94FEE304 pKa = 5.05DD305 pKa = 3.7ATVSYY310 pKa = 10.57VASFDD315 pKa = 4.1DD316 pKa = 5.37LSGEE320 pKa = 3.73NDD322 pKa = 3.52DD323 pKa = 4.92YY324 pKa = 11.74FRR326 pKa = 11.84ITNGTDD332 pKa = 2.73ISATINNVQGSQFFAAQDD350 pKa = 3.56TDD352 pKa = 3.78GVAGNGQSPANLTWSNLNITGLTNINFSAFFAEE385 pKa = 5.45DD386 pKa = 3.79DD387 pKa = 4.12ASDD390 pKa = 4.94AAEE393 pKa = 4.53DD394 pKa = 3.46WDD396 pKa = 3.85NADD399 pKa = 4.36FVDD402 pKa = 3.83VQYY405 pKa = 11.6SFDD408 pKa = 3.43GTTYY412 pKa = 8.02TTFFSIRR419 pKa = 11.84NDD421 pKa = 2.81GSTFNAAPQIDD432 pKa = 4.1TDD434 pKa = 4.32LDD436 pKa = 4.12GVGDD440 pKa = 3.9GAIITDD446 pKa = 3.3NMANYY451 pKa = 9.79SASFPINGTTNPTASTEE468 pKa = 3.47LWIRR472 pKa = 11.84VNVGLDD478 pKa = 3.27SGDD481 pKa = 3.46EE482 pKa = 4.61DD483 pKa = 4.13IAFDD487 pKa = 4.07DD488 pKa = 5.62FIVIGNN494 pKa = 3.95
Molecular weight: 52.92 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A370QFD6|A0A370QFD6_9FLAO Uncharacterized protein (DUF58 family) OS=Marinirhabdus gelatinilytica OX=1703343 GN=C8D94_102258 PE=4 SV=1
MM1 pKa = 7.53IKK3 pKa = 10.29FFRR6 pKa = 11.84HH7 pKa = 4.03IRR9 pKa = 11.84QRR11 pKa = 11.84LIRR14 pKa = 11.84EE15 pKa = 4.01NRR17 pKa = 11.84FSKK20 pKa = 10.73YY21 pKa = 10.02LLYY24 pKa = 10.77AIGEE28 pKa = 4.34IILVVIGILIAEE40 pKa = 4.33NLAPSLKK47 pKa = 10.72GGFSKK52 pKa = 11.05DD53 pKa = 3.34PLRR56 pKa = 5.7
MM1 pKa = 7.53IKK3 pKa = 10.29FFRR6 pKa = 11.84HH7 pKa = 4.03IRR9 pKa = 11.84QRR11 pKa = 11.84LIRR14 pKa = 11.84EE15 pKa = 4.01NRR17 pKa = 11.84FSKK20 pKa = 10.73YY21 pKa = 10.02LLYY24 pKa = 10.77AIGEE28 pKa = 4.34IILVVIGILIAEE40 pKa = 4.33NLAPSLKK47 pKa = 10.72GGFSKK52 pKa = 11.05DD53 pKa = 3.34PLRR56 pKa = 5.7
Molecular weight: 6.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
992534 |
28 |
2692 |
328.4 |
36.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.758 ± 0.034 | 0.744 ± 0.016 |
5.588 ± 0.061 | 6.947 ± 0.043 |
5.176 ± 0.037 | 6.671 ± 0.054 |
1.764 ± 0.022 | 7.258 ± 0.042 |
7.243 ± 0.068 | 9.226 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.192 ± 0.026 | 5.808 ± 0.041 |
3.58 ± 0.031 | 3.633 ± 0.023 |
3.61 ± 0.032 | 6.059 ± 0.034 |
6.245 ± 0.041 | 6.527 ± 0.031 |
1.03 ± 0.016 | 3.941 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
