
Alces alces faeces associated genomovirus MP111
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus lepam1
Average proteome isoelectric point is 7.13
Get precalculated fractions of proteins
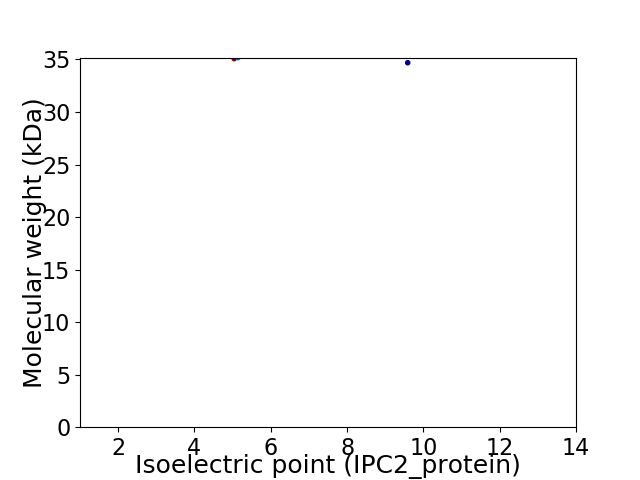
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
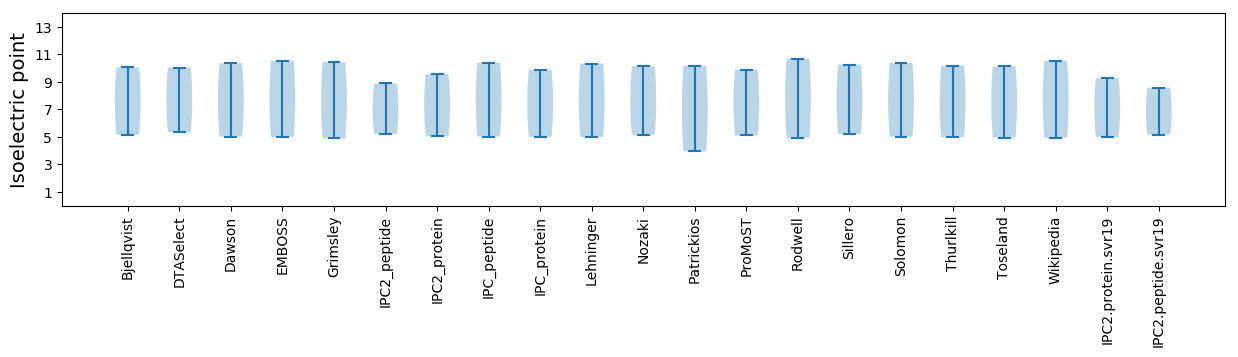
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z5CL71|A0A2Z5CL71_9VIRU Capsid protein OS=Alces alces faeces associated genomovirus MP111 OX=2219111 PE=4 SV=1
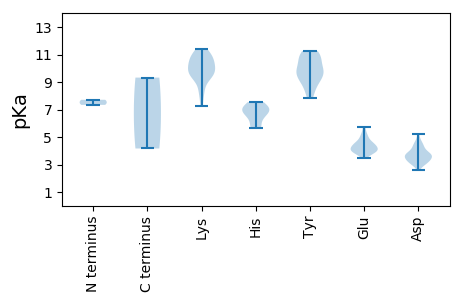
MM1 pKa = 7.71PFSFNARR8 pKa = 11.84YY9 pKa = 9.01FLVTYY14 pKa = 9.29SHH16 pKa = 7.06VDD18 pKa = 3.21EE19 pKa = 5.72LDD21 pKa = 3.3PFAIVDD27 pKa = 3.62HH28 pKa = 6.61FGSLGAEE35 pKa = 3.98CIVARR40 pKa = 11.84EE41 pKa = 4.11RR42 pKa = 11.84YY43 pKa = 9.22ADD45 pKa = 3.72LLGVHH50 pKa = 5.72YY51 pKa = 10.48HH52 pKa = 5.67VFADD56 pKa = 4.64FGRR59 pKa = 11.84KK60 pKa = 8.21FRR62 pKa = 11.84SRR64 pKa = 11.84RR65 pKa = 11.84ANVFDD70 pKa = 3.5VGEE73 pKa = 4.04FHH75 pKa = 7.54PNISPSRR82 pKa = 11.84GTPEE86 pKa = 4.62AGFDD90 pKa = 3.64YY91 pKa = 9.31ATKK94 pKa = 10.77DD95 pKa = 3.16GDD97 pKa = 3.83IVGGGLEE104 pKa = 4.05RR105 pKa = 11.84PSRR108 pKa = 11.84DD109 pKa = 3.09GALSRR114 pKa = 11.84SAKK117 pKa = 8.28WAQIVNAEE125 pKa = 4.27SRR127 pKa = 11.84DD128 pKa = 3.5EE129 pKa = 4.36FYY131 pKa = 10.98RR132 pKa = 11.84LCEE135 pKa = 3.91EE136 pKa = 5.02LDD138 pKa = 3.68PEE140 pKa = 4.71RR141 pKa = 11.84LVCSFGQIQKK151 pKa = 10.02FADD154 pKa = 3.08WKK156 pKa = 10.52FAVTPAPYY164 pKa = 8.43ATPDD168 pKa = 3.53GVFDD172 pKa = 4.28LANYY176 pKa = 9.84GDD178 pKa = 4.11LGEE181 pKa = 4.48SKK183 pKa = 10.86SLILYY188 pKa = 8.74GPSRR192 pKa = 11.84LGKK195 pKa = 7.29TVWSRR200 pKa = 11.84SLGEE204 pKa = 3.86HH205 pKa = 6.9FYY207 pKa = 11.23FGGIFSARR215 pKa = 11.84DD216 pKa = 3.36LDD218 pKa = 4.32RR219 pKa = 11.84DD220 pKa = 3.87DD221 pKa = 3.88VKK223 pKa = 11.41YY224 pKa = 11.03AVFDD228 pKa = 5.22DD229 pKa = 3.44IAGGIKK235 pKa = 10.03FFPRR239 pKa = 11.84FKK241 pKa = 10.79DD242 pKa = 3.12WLGCQMEE249 pKa = 4.46FMVKK253 pKa = 9.27QLYY256 pKa = 9.95RR257 pKa = 11.84DD258 pKa = 3.6PQLFRR263 pKa = 11.84WGRR266 pKa = 11.84PCIWVANTDD275 pKa = 3.19PRR277 pKa = 11.84LDD279 pKa = 3.67MTHH282 pKa = 7.34DD283 pKa = 4.81DD284 pKa = 4.55IMWLEE289 pKa = 4.07ANCIFVEE296 pKa = 3.88ITEE299 pKa = 5.25AIFHH303 pKa = 6.92ANTEE307 pKa = 4.17
MM1 pKa = 7.71PFSFNARR8 pKa = 11.84YY9 pKa = 9.01FLVTYY14 pKa = 9.29SHH16 pKa = 7.06VDD18 pKa = 3.21EE19 pKa = 5.72LDD21 pKa = 3.3PFAIVDD27 pKa = 3.62HH28 pKa = 6.61FGSLGAEE35 pKa = 3.98CIVARR40 pKa = 11.84EE41 pKa = 4.11RR42 pKa = 11.84YY43 pKa = 9.22ADD45 pKa = 3.72LLGVHH50 pKa = 5.72YY51 pKa = 10.48HH52 pKa = 5.67VFADD56 pKa = 4.64FGRR59 pKa = 11.84KK60 pKa = 8.21FRR62 pKa = 11.84SRR64 pKa = 11.84RR65 pKa = 11.84ANVFDD70 pKa = 3.5VGEE73 pKa = 4.04FHH75 pKa = 7.54PNISPSRR82 pKa = 11.84GTPEE86 pKa = 4.62AGFDD90 pKa = 3.64YY91 pKa = 9.31ATKK94 pKa = 10.77DD95 pKa = 3.16GDD97 pKa = 3.83IVGGGLEE104 pKa = 4.05RR105 pKa = 11.84PSRR108 pKa = 11.84DD109 pKa = 3.09GALSRR114 pKa = 11.84SAKK117 pKa = 8.28WAQIVNAEE125 pKa = 4.27SRR127 pKa = 11.84DD128 pKa = 3.5EE129 pKa = 4.36FYY131 pKa = 10.98RR132 pKa = 11.84LCEE135 pKa = 3.91EE136 pKa = 5.02LDD138 pKa = 3.68PEE140 pKa = 4.71RR141 pKa = 11.84LVCSFGQIQKK151 pKa = 10.02FADD154 pKa = 3.08WKK156 pKa = 10.52FAVTPAPYY164 pKa = 8.43ATPDD168 pKa = 3.53GVFDD172 pKa = 4.28LANYY176 pKa = 9.84GDD178 pKa = 4.11LGEE181 pKa = 4.48SKK183 pKa = 10.86SLILYY188 pKa = 8.74GPSRR192 pKa = 11.84LGKK195 pKa = 7.29TVWSRR200 pKa = 11.84SLGEE204 pKa = 3.86HH205 pKa = 6.9FYY207 pKa = 11.23FGGIFSARR215 pKa = 11.84DD216 pKa = 3.36LDD218 pKa = 4.32RR219 pKa = 11.84DD220 pKa = 3.87DD221 pKa = 3.88VKK223 pKa = 11.41YY224 pKa = 11.03AVFDD228 pKa = 5.22DD229 pKa = 3.44IAGGIKK235 pKa = 10.03FFPRR239 pKa = 11.84FKK241 pKa = 10.79DD242 pKa = 3.12WLGCQMEE249 pKa = 4.46FMVKK253 pKa = 9.27QLYY256 pKa = 9.95RR257 pKa = 11.84DD258 pKa = 3.6PQLFRR263 pKa = 11.84WGRR266 pKa = 11.84PCIWVANTDD275 pKa = 3.19PRR277 pKa = 11.84LDD279 pKa = 3.67MTHH282 pKa = 7.34DD283 pKa = 4.81DD284 pKa = 4.55IMWLEE289 pKa = 4.07ANCIFVEE296 pKa = 3.88ITEE299 pKa = 5.25AIFHH303 pKa = 6.92ANTEE307 pKa = 4.17
Molecular weight: 35.08 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z5CL71|A0A2Z5CL71_9VIRU Capsid protein OS=Alces alces faeces associated genomovirus MP111 OX=2219111 PE=4 SV=1
MM1 pKa = 7.34ARR3 pKa = 11.84TWASIYY9 pKa = 10.05RR10 pKa = 11.84KK11 pKa = 8.88PFRR14 pKa = 11.84RR15 pKa = 11.84TRR17 pKa = 11.84PVRR20 pKa = 11.84GGRR23 pKa = 11.84TTRR26 pKa = 11.84RR27 pKa = 11.84STKK30 pKa = 9.79KK31 pKa = 9.68RR32 pKa = 11.84PTYY35 pKa = 9.62RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 10.23SRR40 pKa = 11.84TMSKK44 pKa = 10.37KK45 pKa = 10.11RR46 pKa = 11.84ILNISSRR53 pKa = 11.84KK54 pKa = 9.6KK55 pKa = 9.61RR56 pKa = 11.84DD57 pKa = 2.93TMAPIQQNGSSGTPAIGALNINGAPGNPGNATFVFWCATARR98 pKa = 11.84DD99 pKa = 3.88NTVAQGGAASSVYY112 pKa = 10.56NPEE115 pKa = 3.46TRR117 pKa = 11.84TATTCYY123 pKa = 9.78MRR125 pKa = 11.84GLKK128 pKa = 10.33EE129 pKa = 3.93NVEE132 pKa = 4.48MQTSTGIPWQWRR144 pKa = 11.84RR145 pKa = 11.84ICFTYY150 pKa = 11.02KK151 pKa = 11.0DD152 pKa = 3.83MTEE155 pKa = 3.89VCATNNGGISGFYY168 pKa = 9.85EE169 pKa = 4.29GSNGYY174 pKa = 9.05GRR176 pKa = 11.84LMYY179 pKa = 11.25NMFSSGSAADD189 pKa = 3.65VKK191 pKa = 10.97AYY193 pKa = 7.81TALQTLLFRR202 pKa = 11.84GTIGVDD208 pKa = 2.58WNDD211 pKa = 4.22YY212 pKa = 8.15ITAPIDD218 pKa = 3.54TQRR221 pKa = 11.84VGLKK225 pKa = 9.45YY226 pKa = 10.83DD227 pKa = 3.31KK228 pKa = 10.94TRR230 pKa = 11.84VIQSGNANGMLRR242 pKa = 11.84KK243 pKa = 9.46YY244 pKa = 10.35QMWHH248 pKa = 6.4PMNKK252 pKa = 9.54NLVYY256 pKa = 10.62DD257 pKa = 4.29DD258 pKa = 4.47SEE260 pKa = 5.04KK261 pKa = 11.28GGADD265 pKa = 2.99TAGYY269 pKa = 9.69FSIGGKK275 pKa = 9.49PGMGDD280 pKa = 3.73FLVLDD285 pKa = 3.86IFMPGKK291 pKa = 10.31GSSSSDD297 pKa = 3.27VLSFEE302 pKa = 4.53ATSTLYY308 pKa = 9.64WHH310 pKa = 7.08EE311 pKa = 4.2KK312 pKa = 9.32
MM1 pKa = 7.34ARR3 pKa = 11.84TWASIYY9 pKa = 10.05RR10 pKa = 11.84KK11 pKa = 8.88PFRR14 pKa = 11.84RR15 pKa = 11.84TRR17 pKa = 11.84PVRR20 pKa = 11.84GGRR23 pKa = 11.84TTRR26 pKa = 11.84RR27 pKa = 11.84STKK30 pKa = 9.79KK31 pKa = 9.68RR32 pKa = 11.84PTYY35 pKa = 9.62RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 10.23SRR40 pKa = 11.84TMSKK44 pKa = 10.37KK45 pKa = 10.11RR46 pKa = 11.84ILNISSRR53 pKa = 11.84KK54 pKa = 9.6KK55 pKa = 9.61RR56 pKa = 11.84DD57 pKa = 2.93TMAPIQQNGSSGTPAIGALNINGAPGNPGNATFVFWCATARR98 pKa = 11.84DD99 pKa = 3.88NTVAQGGAASSVYY112 pKa = 10.56NPEE115 pKa = 3.46TRR117 pKa = 11.84TATTCYY123 pKa = 9.78MRR125 pKa = 11.84GLKK128 pKa = 10.33EE129 pKa = 3.93NVEE132 pKa = 4.48MQTSTGIPWQWRR144 pKa = 11.84RR145 pKa = 11.84ICFTYY150 pKa = 11.02KK151 pKa = 11.0DD152 pKa = 3.83MTEE155 pKa = 3.89VCATNNGGISGFYY168 pKa = 9.85EE169 pKa = 4.29GSNGYY174 pKa = 9.05GRR176 pKa = 11.84LMYY179 pKa = 11.25NMFSSGSAADD189 pKa = 3.65VKK191 pKa = 10.97AYY193 pKa = 7.81TALQTLLFRR202 pKa = 11.84GTIGVDD208 pKa = 2.58WNDD211 pKa = 4.22YY212 pKa = 8.15ITAPIDD218 pKa = 3.54TQRR221 pKa = 11.84VGLKK225 pKa = 9.45YY226 pKa = 10.83DD227 pKa = 3.31KK228 pKa = 10.94TRR230 pKa = 11.84VIQSGNANGMLRR242 pKa = 11.84KK243 pKa = 9.46YY244 pKa = 10.35QMWHH248 pKa = 6.4PMNKK252 pKa = 9.54NLVYY256 pKa = 10.62DD257 pKa = 4.29DD258 pKa = 4.47SEE260 pKa = 5.04KK261 pKa = 11.28GGADD265 pKa = 2.99TAGYY269 pKa = 9.69FSIGGKK275 pKa = 9.49PGMGDD280 pKa = 3.73FLVLDD285 pKa = 3.86IFMPGKK291 pKa = 10.31GSSSSDD297 pKa = 3.27VLSFEE302 pKa = 4.53ATSTLYY308 pKa = 9.64WHH310 pKa = 7.08EE311 pKa = 4.2KK312 pKa = 9.32
Molecular weight: 34.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
619 |
307 |
312 |
309.5 |
34.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.754 ± 0.291 | 1.616 ± 0.254 |
6.785 ± 1.748 | 4.2 ± 1.245 |
6.139 ± 1.988 | 9.37 ± 0.918 |
1.616 ± 0.741 | 4.847 ± 0.03 |
4.847 ± 0.946 | 5.816 ± 1.011 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.908 ± 0.958 | 4.2 ± 1.193 |
4.523 ± 0.271 | 2.423 ± 0.351 |
7.916 ± 0.317 | 6.785 ± 0.934 |
6.462 ± 2.399 | 5.17 ± 0.763 |
2.262 ± 0.014 | 4.362 ± 0.339 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
