
Synechococcus phage S-CAM8
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; Acionnavirus; unclassified Acionnavirus
Average proteome isoelectric point is 5.63
Get precalculated fractions of proteins
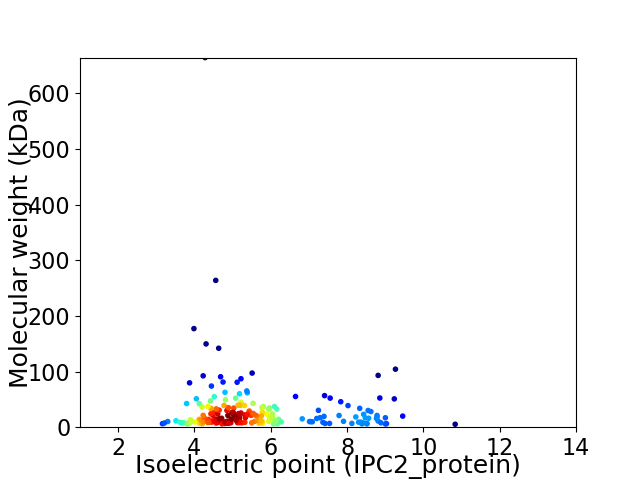
Virtual 2D-PAGE plot for 204 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions



Protein with the lowest isoelectric point:
>tr|G8EXL5|G8EXL5_9CAUD Uncharacterized protein OS=Synechococcus phage S-CAM8 OX=754038 GN=SXCG_00168 PE=4 SV=1
MM1 pKa = 7.74AYY3 pKa = 9.73YY4 pKa = 10.45YY5 pKa = 9.97PEE7 pKa = 5.61GYY9 pKa = 10.04FGPICDD15 pKa = 4.56TPLSDD20 pKa = 5.45DD21 pKa = 4.43QISSQQRR28 pKa = 11.84DD29 pKa = 3.5TPAFDD34 pKa = 3.28EE35 pKa = 4.86GEE37 pKa = 4.12EE38 pKa = 4.09EE39 pKa = 4.72RR40 pKa = 11.84VQNYY44 pKa = 8.04PAEE47 pKa = 4.58GNSDD51 pKa = 3.59IPWWFAYY58 pKa = 9.83KK59 pKa = 10.51DD60 pKa = 3.41ITGDD64 pKa = 3.59LPSVLISTRR73 pKa = 11.84CKK75 pKa = 9.97QRR77 pKa = 11.84EE78 pKa = 4.07DD79 pKa = 3.65GSFFDD84 pKa = 5.59CEE86 pKa = 4.03YY87 pKa = 10.48EE88 pKa = 4.17YY89 pKa = 11.24ASPVDD94 pKa = 4.07FPEE97 pKa = 4.63INVPQEE103 pKa = 3.88DD104 pKa = 4.01FGLGEE109 pKa = 4.0QFFVPVITPEE119 pKa = 3.9SCSPFDD125 pKa = 3.95ADD127 pKa = 5.3INILPISFITPSGGLVTKK145 pKa = 9.92YY146 pKa = 10.22KK147 pKa = 10.27KK148 pKa = 10.16EE149 pKa = 3.8KK150 pKa = 10.2SSPVTYY156 pKa = 10.51AVTSTRR162 pKa = 11.84GFTEE166 pKa = 4.01VQSDD170 pKa = 4.13LDD172 pKa = 3.57VSFNDD177 pKa = 3.77EE178 pKa = 3.92GTQVIVTGTGSAIVTFEE195 pKa = 4.83LRR197 pKa = 11.84WDD199 pKa = 4.41DD200 pKa = 4.9DD201 pKa = 4.15PGTSGLAVGTLTIEE215 pKa = 4.48GQSVSQSGEE224 pKa = 3.93EE225 pKa = 4.4GSTAMSVQLSSGTYY239 pKa = 8.82PISLSGEE246 pKa = 4.1SSGSGSRR253 pKa = 11.84TRR255 pKa = 11.84NNNKK259 pKa = 9.1TIEE262 pKa = 4.13YY263 pKa = 10.28DD264 pKa = 3.9DD265 pKa = 6.44DD266 pKa = 4.29IDD268 pKa = 4.79EE269 pKa = 6.06DD270 pKa = 4.42GFDD273 pKa = 3.85VNATLSIQSVDD284 pKa = 4.03NIDD287 pKa = 4.68GIQIDD292 pKa = 4.64SFWSDD297 pKa = 2.73EE298 pKa = 4.03GNAYY302 pKa = 9.94AVWTNPEE309 pKa = 3.97TCTLPRR315 pKa = 11.84EE316 pKa = 4.4EE317 pKa = 4.56QDD319 pKa = 3.15VTYY322 pKa = 10.38KK323 pKa = 10.56IPIPTAGTYY332 pKa = 10.84GFTFASDD339 pKa = 3.4DD340 pKa = 3.76GGTVIINEE348 pKa = 4.03NDD350 pKa = 3.71TIFNNLPGGIFKK362 pKa = 10.65SGSLSTPYY370 pKa = 10.55SATRR374 pKa = 11.84NLDD377 pKa = 3.31AGILEE382 pKa = 4.21MTVRR386 pKa = 11.84CTNSDD391 pKa = 3.06AGFTTDD397 pKa = 4.94GEE399 pKa = 4.48PSGLAYY405 pKa = 10.25AWWRR409 pKa = 11.84NPGGWYY415 pKa = 9.82IKK417 pKa = 9.19ICKK420 pKa = 10.07GGTCAGEE427 pKa = 4.32TIVPWVRR434 pKa = 11.84SGPHH438 pKa = 4.84QLWSEE443 pKa = 3.78FMNNYY448 pKa = 9.73AVYY451 pKa = 9.59TSNTEE456 pKa = 3.9PSLGTHH462 pKa = 4.94TTSYY466 pKa = 10.16TIYY469 pKa = 10.05IDD471 pKa = 4.67EE472 pKa = 4.56EE473 pKa = 4.38DD474 pKa = 4.62DD475 pKa = 4.19YY476 pKa = 11.47LLEE479 pKa = 4.2MQADD483 pKa = 4.05NNGTISWDD491 pKa = 3.24GTQVLTSSSYY501 pKa = 7.71TTSSTYY507 pKa = 10.0TIPNVSVGPHH517 pKa = 5.08TLSVSVTNVANGGTDD532 pKa = 2.92IDD534 pKa = 3.69TWGNNPGGIAWTLTQPSDD552 pKa = 3.34TVTTTEE558 pKa = 4.2TVTTEE563 pKa = 4.15TPNEE567 pKa = 3.71IQATFNSDD575 pKa = 2.35GDD577 pKa = 3.9IVVTGFGTGRR587 pKa = 11.84IQLIFEE593 pKa = 4.47WDD595 pKa = 3.8DD596 pKa = 3.42DD597 pKa = 4.23PNRR600 pKa = 11.84YY601 pKa = 7.52GTALGSIEE609 pKa = 4.05IAGKK613 pKa = 8.31TFVQTTGKK621 pKa = 10.07EE622 pKa = 4.0RR623 pKa = 11.84GNDD626 pKa = 3.13SYY628 pKa = 11.75TFTARR633 pKa = 11.84AGRR636 pKa = 11.84TYY638 pKa = 10.51TAVIEE643 pKa = 4.5DD644 pKa = 4.02NPSGFTRR651 pKa = 11.84KK652 pKa = 9.64RR653 pKa = 11.84SNTRR657 pKa = 11.84LCFFDD662 pKa = 6.14SDD664 pKa = 5.82DD665 pKa = 4.59DD666 pKa = 4.18DD667 pKa = 6.04CNAKK671 pKa = 10.28LDD673 pKa = 4.05IASVTYY679 pKa = 10.44SGTVSSSSTDD689 pKa = 3.21TNTTTIEE696 pKa = 3.68EE697 pKa = 4.41SIIARR702 pKa = 11.84STDD705 pKa = 3.6LNIGSNIDD713 pKa = 3.44NNNLIWHH720 pKa = 6.11TRR722 pKa = 11.84LATGYY727 pKa = 9.91EE728 pKa = 4.27YY729 pKa = 10.28STTQQ733 pKa = 2.97
MM1 pKa = 7.74AYY3 pKa = 9.73YY4 pKa = 10.45YY5 pKa = 9.97PEE7 pKa = 5.61GYY9 pKa = 10.04FGPICDD15 pKa = 4.56TPLSDD20 pKa = 5.45DD21 pKa = 4.43QISSQQRR28 pKa = 11.84DD29 pKa = 3.5TPAFDD34 pKa = 3.28EE35 pKa = 4.86GEE37 pKa = 4.12EE38 pKa = 4.09EE39 pKa = 4.72RR40 pKa = 11.84VQNYY44 pKa = 8.04PAEE47 pKa = 4.58GNSDD51 pKa = 3.59IPWWFAYY58 pKa = 9.83KK59 pKa = 10.51DD60 pKa = 3.41ITGDD64 pKa = 3.59LPSVLISTRR73 pKa = 11.84CKK75 pKa = 9.97QRR77 pKa = 11.84EE78 pKa = 4.07DD79 pKa = 3.65GSFFDD84 pKa = 5.59CEE86 pKa = 4.03YY87 pKa = 10.48EE88 pKa = 4.17YY89 pKa = 11.24ASPVDD94 pKa = 4.07FPEE97 pKa = 4.63INVPQEE103 pKa = 3.88DD104 pKa = 4.01FGLGEE109 pKa = 4.0QFFVPVITPEE119 pKa = 3.9SCSPFDD125 pKa = 3.95ADD127 pKa = 5.3INILPISFITPSGGLVTKK145 pKa = 9.92YY146 pKa = 10.22KK147 pKa = 10.27KK148 pKa = 10.16EE149 pKa = 3.8KK150 pKa = 10.2SSPVTYY156 pKa = 10.51AVTSTRR162 pKa = 11.84GFTEE166 pKa = 4.01VQSDD170 pKa = 4.13LDD172 pKa = 3.57VSFNDD177 pKa = 3.77EE178 pKa = 3.92GTQVIVTGTGSAIVTFEE195 pKa = 4.83LRR197 pKa = 11.84WDD199 pKa = 4.41DD200 pKa = 4.9DD201 pKa = 4.15PGTSGLAVGTLTIEE215 pKa = 4.48GQSVSQSGEE224 pKa = 3.93EE225 pKa = 4.4GSTAMSVQLSSGTYY239 pKa = 8.82PISLSGEE246 pKa = 4.1SSGSGSRR253 pKa = 11.84TRR255 pKa = 11.84NNNKK259 pKa = 9.1TIEE262 pKa = 4.13YY263 pKa = 10.28DD264 pKa = 3.9DD265 pKa = 6.44DD266 pKa = 4.29IDD268 pKa = 4.79EE269 pKa = 6.06DD270 pKa = 4.42GFDD273 pKa = 3.85VNATLSIQSVDD284 pKa = 4.03NIDD287 pKa = 4.68GIQIDD292 pKa = 4.64SFWSDD297 pKa = 2.73EE298 pKa = 4.03GNAYY302 pKa = 9.94AVWTNPEE309 pKa = 3.97TCTLPRR315 pKa = 11.84EE316 pKa = 4.4EE317 pKa = 4.56QDD319 pKa = 3.15VTYY322 pKa = 10.38KK323 pKa = 10.56IPIPTAGTYY332 pKa = 10.84GFTFASDD339 pKa = 3.4DD340 pKa = 3.76GGTVIINEE348 pKa = 4.03NDD350 pKa = 3.71TIFNNLPGGIFKK362 pKa = 10.65SGSLSTPYY370 pKa = 10.55SATRR374 pKa = 11.84NLDD377 pKa = 3.31AGILEE382 pKa = 4.21MTVRR386 pKa = 11.84CTNSDD391 pKa = 3.06AGFTTDD397 pKa = 4.94GEE399 pKa = 4.48PSGLAYY405 pKa = 10.25AWWRR409 pKa = 11.84NPGGWYY415 pKa = 9.82IKK417 pKa = 9.19ICKK420 pKa = 10.07GGTCAGEE427 pKa = 4.32TIVPWVRR434 pKa = 11.84SGPHH438 pKa = 4.84QLWSEE443 pKa = 3.78FMNNYY448 pKa = 9.73AVYY451 pKa = 9.59TSNTEE456 pKa = 3.9PSLGTHH462 pKa = 4.94TTSYY466 pKa = 10.16TIYY469 pKa = 10.05IDD471 pKa = 4.67EE472 pKa = 4.56EE473 pKa = 4.38DD474 pKa = 4.62DD475 pKa = 4.19YY476 pKa = 11.47LLEE479 pKa = 4.2MQADD483 pKa = 4.05NNGTISWDD491 pKa = 3.24GTQVLTSSSYY501 pKa = 7.71TTSSTYY507 pKa = 10.0TIPNVSVGPHH517 pKa = 5.08TLSVSVTNVANGGTDD532 pKa = 2.92IDD534 pKa = 3.69TWGNNPGGIAWTLTQPSDD552 pKa = 3.34TVTTTEE558 pKa = 4.2TVTTEE563 pKa = 4.15TPNEE567 pKa = 3.71IQATFNSDD575 pKa = 2.35GDD577 pKa = 3.9IVVTGFGTGRR587 pKa = 11.84IQLIFEE593 pKa = 4.47WDD595 pKa = 3.8DD596 pKa = 3.42DD597 pKa = 4.23PNRR600 pKa = 11.84YY601 pKa = 7.52GTALGSIEE609 pKa = 4.05IAGKK613 pKa = 8.31TFVQTTGKK621 pKa = 10.07EE622 pKa = 4.0RR623 pKa = 11.84GNDD626 pKa = 3.13SYY628 pKa = 11.75TFTARR633 pKa = 11.84AGRR636 pKa = 11.84TYY638 pKa = 10.51TAVIEE643 pKa = 4.5DD644 pKa = 4.02NPSGFTRR651 pKa = 11.84KK652 pKa = 9.64RR653 pKa = 11.84SNTRR657 pKa = 11.84LCFFDD662 pKa = 6.14SDD664 pKa = 5.82DD665 pKa = 4.59DD666 pKa = 4.18DD667 pKa = 6.04CNAKK671 pKa = 10.28LDD673 pKa = 4.05IASVTYY679 pKa = 10.44SGTVSSSSTDD689 pKa = 3.21TNTTTIEE696 pKa = 3.68EE697 pKa = 4.41SIIARR702 pKa = 11.84STDD705 pKa = 3.6LNIGSNIDD713 pKa = 3.44NNNLIWHH720 pKa = 6.11TRR722 pKa = 11.84LATGYY727 pKa = 9.91EE728 pKa = 4.27YY729 pKa = 10.28STTQQ733 pKa = 2.97
Molecular weight: 79.76 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G8EXQ7|G8EXQ7_9CAUD Uncharacterized protein OS=Synechococcus phage S-CAM8 OX=754038 GN=SXCG_00008 PE=4 SV=1
MM1 pKa = 7.52AVRR4 pKa = 11.84SKK6 pKa = 11.0SLSGGNLIQSKK17 pKa = 9.43PKK19 pKa = 10.2KK20 pKa = 9.5SRR22 pKa = 11.84QGSGQHH28 pKa = 5.15TKK30 pKa = 10.34YY31 pKa = 10.86ASSSRR36 pKa = 11.84NNAKK40 pKa = 9.85KK41 pKa = 10.23RR42 pKa = 11.84YY43 pKa = 8.5RR44 pKa = 11.84GQGRR48 pKa = 3.34
MM1 pKa = 7.52AVRR4 pKa = 11.84SKK6 pKa = 11.0SLSGGNLIQSKK17 pKa = 9.43PKK19 pKa = 10.2KK20 pKa = 9.5SRR22 pKa = 11.84QGSGQHH28 pKa = 5.15TKK30 pKa = 10.34YY31 pKa = 10.86ASSSRR36 pKa = 11.84NNAKK40 pKa = 9.85KK41 pKa = 10.23RR42 pKa = 11.84YY43 pKa = 8.5RR44 pKa = 11.84GQGRR48 pKa = 3.34
Molecular weight: 5.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
54297 |
48 |
6167 |
266.2 |
29.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.89 ± 0.215 | 0.88 ± 0.099 |
6.68 ± 0.135 | 6.166 ± 0.242 |
4.221 ± 0.114 | 7.918 ± 0.31 |
1.37 ± 0.116 | 6.435 ± 0.19 |
5.709 ± 0.417 | 7.22 ± 0.132 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.089 ± 0.227 | 5.785 ± 0.176 |
3.825 ± 0.137 | 3.761 ± 0.105 |
3.939 ± 0.146 | 7.43 ± 0.292 |
7.638 ± 0.411 | 6.632 ± 0.219 |
1.147 ± 0.081 | 4.265 ± 0.138 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
