
Salisediminibacterium haloalkalitolerans
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Salisediminibacterium
Average proteome isoelectric point is 5.69
Get precalculated fractions of proteins
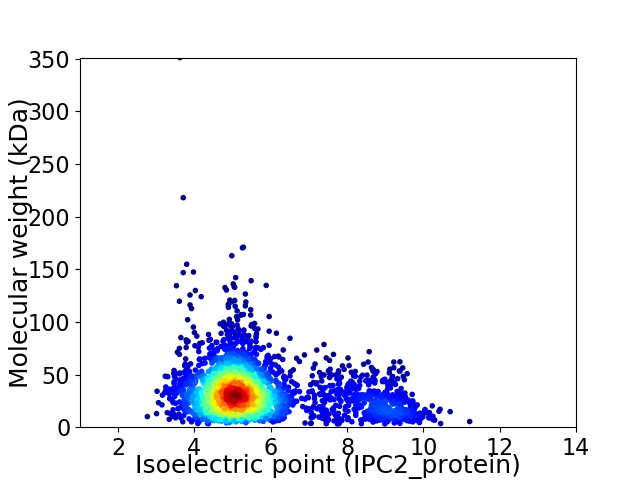
Virtual 2D-PAGE plot for 2769 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H9VJ15|A0A1H9VJ15_9BACI Uncharacterized protein OS=Salisediminibacterium haloalkalitolerans OX=1464123 GN=SAMN05444126_12118 PE=4 SV=1
MM1 pKa = 7.53NIYY4 pKa = 9.76GKK6 pKa = 10.15KK7 pKa = 9.71VLVAGSLLLLAACGNDD23 pKa = 3.37EE24 pKa = 4.15NEE26 pKa = 3.99EE27 pKa = 3.76LAAEE31 pKa = 4.2NEE33 pKa = 3.79ADD35 pKa = 3.7EE36 pKa = 4.42NNRR39 pKa = 11.84GILSDD44 pKa = 4.35DD45 pKa = 4.05NEE47 pKa = 4.34NLADD51 pKa = 4.65DD52 pKa = 4.87NNEE55 pKa = 3.66QTEE58 pKa = 4.17NSDD61 pKa = 3.62SKK63 pKa = 11.52EE64 pKa = 4.05NAPEE68 pKa = 4.38PDD70 pKa = 3.45PEE72 pKa = 4.12EE73 pKa = 4.44NIGNFNGNLAQGGRR87 pKa = 11.84QAYY90 pKa = 9.66HH91 pKa = 6.64EE92 pKa = 4.38DD93 pKa = 3.4VFYY96 pKa = 11.43VSAALDD102 pKa = 3.94EE103 pKa = 4.27EE104 pKa = 4.5HH105 pKa = 7.24GIYY108 pKa = 10.36AVPLEE113 pKa = 4.34TNEE116 pKa = 3.78YY117 pKa = 9.69EE118 pKa = 4.31QLTDD122 pKa = 4.37GKK124 pKa = 10.11AQDD127 pKa = 3.43MHH129 pKa = 5.96VTDD132 pKa = 4.78DD133 pKa = 4.3GIYY136 pKa = 9.48YY137 pKa = 8.83LTDD140 pKa = 3.73PGYY143 pKa = 11.13DD144 pKa = 3.41NDD146 pKa = 3.8SDD148 pKa = 4.24EE149 pKa = 4.96IKK151 pKa = 10.38LIHH154 pKa = 6.25YY155 pKa = 10.23AFDD158 pKa = 3.72TDD160 pKa = 3.82EE161 pKa = 5.51KK162 pKa = 10.73NTLDD166 pKa = 4.02EE167 pKa = 4.81GNLRR171 pKa = 11.84NLQMNHH177 pKa = 6.89DD178 pKa = 4.22GLWYY182 pKa = 10.49LKK184 pKa = 10.45AGNEE188 pKa = 3.87GGMNFINKK196 pKa = 9.07NMEE199 pKa = 3.83TGEE202 pKa = 4.0FDD204 pKa = 3.66EE205 pKa = 5.54FSSGQSQIIIDD216 pKa = 4.06GDD218 pKa = 3.91DD219 pKa = 4.74AISQAEE225 pKa = 4.17TFIYY229 pKa = 10.92DD230 pKa = 3.14MDD232 pKa = 5.65GPDD235 pKa = 3.57GTIEE239 pKa = 4.33DD240 pKa = 4.17EE241 pKa = 4.35PVVEE245 pKa = 4.33NATPPVILKK254 pKa = 10.59DD255 pKa = 3.26RR256 pKa = 11.84VVYY259 pKa = 10.55YY260 pKa = 10.3NGPNGIEE267 pKa = 3.94AHH269 pKa = 6.79DD270 pKa = 4.83LDD272 pKa = 4.47SGQTIKK278 pKa = 9.41ITNEE282 pKa = 3.49RR283 pKa = 11.84VSDD286 pKa = 3.74FNVRR290 pKa = 11.84NGIVYY295 pKa = 7.33FTPPFQSEE303 pKa = 4.23AARR306 pKa = 11.84EE307 pKa = 4.26SYY309 pKa = 10.52RR310 pKa = 11.84VDD312 pKa = 3.28ADD314 pKa = 3.52GTNEE318 pKa = 3.86EE319 pKa = 4.15NLEE322 pKa = 4.58GISYY326 pKa = 8.5TYY328 pKa = 11.81YY329 pKa = 10.23MFDD332 pKa = 3.58DD333 pKa = 4.67FAVHH337 pKa = 5.85SSGRR341 pKa = 11.84HH342 pKa = 3.74GVILYY347 pKa = 10.55SKK349 pKa = 8.7ITYY352 pKa = 10.09DD353 pKa = 3.42SEE355 pKa = 4.22EE356 pKa = 4.25SEE358 pKa = 4.33KK359 pKa = 10.34IYY361 pKa = 9.57YY362 pKa = 7.41TNPGMEE368 pKa = 4.44DD369 pKa = 2.91
MM1 pKa = 7.53NIYY4 pKa = 9.76GKK6 pKa = 10.15KK7 pKa = 9.71VLVAGSLLLLAACGNDD23 pKa = 3.37EE24 pKa = 4.15NEE26 pKa = 3.99EE27 pKa = 3.76LAAEE31 pKa = 4.2NEE33 pKa = 3.79ADD35 pKa = 3.7EE36 pKa = 4.42NNRR39 pKa = 11.84GILSDD44 pKa = 4.35DD45 pKa = 4.05NEE47 pKa = 4.34NLADD51 pKa = 4.65DD52 pKa = 4.87NNEE55 pKa = 3.66QTEE58 pKa = 4.17NSDD61 pKa = 3.62SKK63 pKa = 11.52EE64 pKa = 4.05NAPEE68 pKa = 4.38PDD70 pKa = 3.45PEE72 pKa = 4.12EE73 pKa = 4.44NIGNFNGNLAQGGRR87 pKa = 11.84QAYY90 pKa = 9.66HH91 pKa = 6.64EE92 pKa = 4.38DD93 pKa = 3.4VFYY96 pKa = 11.43VSAALDD102 pKa = 3.94EE103 pKa = 4.27EE104 pKa = 4.5HH105 pKa = 7.24GIYY108 pKa = 10.36AVPLEE113 pKa = 4.34TNEE116 pKa = 3.78YY117 pKa = 9.69EE118 pKa = 4.31QLTDD122 pKa = 4.37GKK124 pKa = 10.11AQDD127 pKa = 3.43MHH129 pKa = 5.96VTDD132 pKa = 4.78DD133 pKa = 4.3GIYY136 pKa = 9.48YY137 pKa = 8.83LTDD140 pKa = 3.73PGYY143 pKa = 11.13DD144 pKa = 3.41NDD146 pKa = 3.8SDD148 pKa = 4.24EE149 pKa = 4.96IKK151 pKa = 10.38LIHH154 pKa = 6.25YY155 pKa = 10.23AFDD158 pKa = 3.72TDD160 pKa = 3.82EE161 pKa = 5.51KK162 pKa = 10.73NTLDD166 pKa = 4.02EE167 pKa = 4.81GNLRR171 pKa = 11.84NLQMNHH177 pKa = 6.89DD178 pKa = 4.22GLWYY182 pKa = 10.49LKK184 pKa = 10.45AGNEE188 pKa = 3.87GGMNFINKK196 pKa = 9.07NMEE199 pKa = 3.83TGEE202 pKa = 4.0FDD204 pKa = 3.66EE205 pKa = 5.54FSSGQSQIIIDD216 pKa = 4.06GDD218 pKa = 3.91DD219 pKa = 4.74AISQAEE225 pKa = 4.17TFIYY229 pKa = 10.92DD230 pKa = 3.14MDD232 pKa = 5.65GPDD235 pKa = 3.57GTIEE239 pKa = 4.33DD240 pKa = 4.17EE241 pKa = 4.35PVVEE245 pKa = 4.33NATPPVILKK254 pKa = 10.59DD255 pKa = 3.26RR256 pKa = 11.84VVYY259 pKa = 10.55YY260 pKa = 10.3NGPNGIEE267 pKa = 3.94AHH269 pKa = 6.79DD270 pKa = 4.83LDD272 pKa = 4.47SGQTIKK278 pKa = 9.41ITNEE282 pKa = 3.49RR283 pKa = 11.84VSDD286 pKa = 3.74FNVRR290 pKa = 11.84NGIVYY295 pKa = 7.33FTPPFQSEE303 pKa = 4.23AARR306 pKa = 11.84EE307 pKa = 4.26SYY309 pKa = 10.52RR310 pKa = 11.84VDD312 pKa = 3.28ADD314 pKa = 3.52GTNEE318 pKa = 3.86EE319 pKa = 4.15NLEE322 pKa = 4.58GISYY326 pKa = 8.5TYY328 pKa = 11.81YY329 pKa = 10.23MFDD332 pKa = 3.58DD333 pKa = 4.67FAVHH337 pKa = 5.85SSGRR341 pKa = 11.84HH342 pKa = 3.74GVILYY347 pKa = 10.55SKK349 pKa = 8.7ITYY352 pKa = 10.09DD353 pKa = 3.42SEE355 pKa = 4.22EE356 pKa = 4.25SEE358 pKa = 4.33KK359 pKa = 10.34IYY361 pKa = 9.57YY362 pKa = 7.41TNPGMEE368 pKa = 4.44DD369 pKa = 2.91
Molecular weight: 41.24 kDa
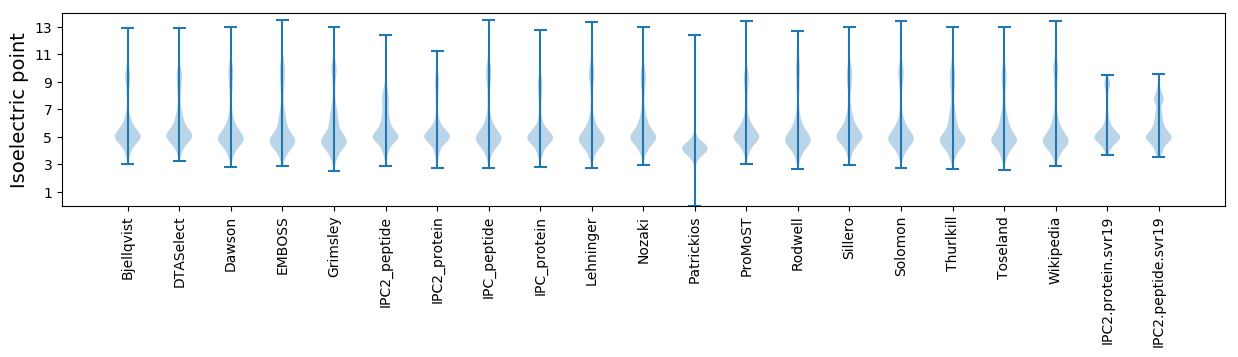
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H9WPY8|A0A1H9WPY8_9BACI Sucrose-6-phosphate hydrolase OS=Salisediminibacterium haloalkalitolerans OX=1464123 GN=SAMN05444126_1424 PE=3 SV=1
MM1 pKa = 7.51SKK3 pKa = 9.27ATFQPNNRR11 pKa = 11.84KK12 pKa = 9.23RR13 pKa = 11.84KK14 pKa = 8.22KK15 pKa = 8.69VHH17 pKa = 5.45GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.29NGRR29 pKa = 11.84QVLQRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.93GRR40 pKa = 11.84KK41 pKa = 8.7VLSAA45 pKa = 4.05
MM1 pKa = 7.51SKK3 pKa = 9.27ATFQPNNRR11 pKa = 11.84KK12 pKa = 9.23RR13 pKa = 11.84KK14 pKa = 8.22KK15 pKa = 8.69VHH17 pKa = 5.45GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.29NGRR29 pKa = 11.84QVLQRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.93GRR40 pKa = 11.84KK41 pKa = 8.7VLSAA45 pKa = 4.05
Molecular weight: 5.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
857196 |
27 |
3116 |
309.6 |
34.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.482 ± 0.054 | 0.623 ± 0.014 |
6.019 ± 0.056 | 8.353 ± 0.07 |
4.322 ± 0.04 | 7.172 ± 0.045 |
2.205 ± 0.025 | 6.598 ± 0.043 |
5.211 ± 0.049 | 9.256 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.93 ± 0.024 | 3.914 ± 0.034 |
3.816 ± 0.024 | 3.721 ± 0.033 |
4.658 ± 0.035 | 5.778 ± 0.033 |
5.575 ± 0.03 | 7.057 ± 0.04 |
1.0 ± 0.016 | 3.307 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
