
Serinibacter arcticus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Beutenbergiaceae; Serinibacter
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins
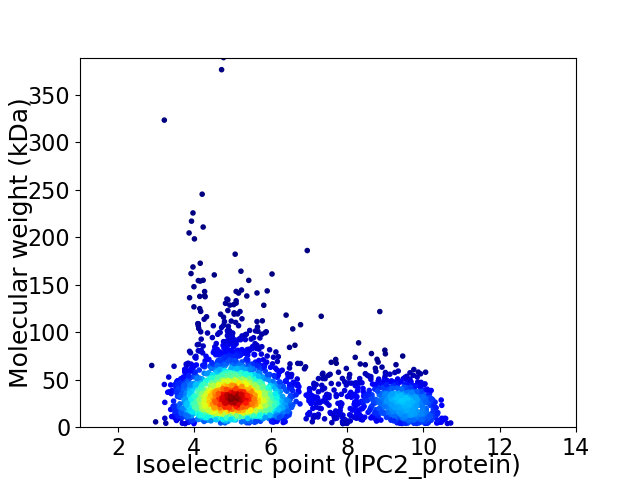
Virtual 2D-PAGE plot for 2999 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Z1DXW7|A0A4Z1DXW7_9MICO Phosphate starvation-inducible protein PhoH predicted ATPase OS=Serinibacter arcticus OX=1655435 GN=SERN_1969 PE=4 SV=1
MM1 pKa = 7.39AVVGLAASSLVLAGCSSEE19 pKa = 4.22RR20 pKa = 11.84GEE22 pKa = 4.49GGSDD26 pKa = 3.2GSSSEE31 pKa = 4.25GGSGTSSEE39 pKa = 4.35TGEE42 pKa = 4.16AAGPTTFTFASSSDD56 pKa = 3.77PASLDD61 pKa = 3.26PAFASDD67 pKa = 3.92GEE69 pKa = 4.5SFRR72 pKa = 11.84VARR75 pKa = 11.84QMFEE79 pKa = 3.81GLVGVEE85 pKa = 4.84PGTADD90 pKa = 3.86PAPLLAEE97 pKa = 4.14SWDD100 pKa = 3.68VSDD103 pKa = 6.37DD104 pKa = 3.4GLEE107 pKa = 3.93YY108 pKa = 10.56TFQLKK113 pKa = 10.46EE114 pKa = 4.0GVSFHH119 pKa = 7.64DD120 pKa = 3.56GTPFNGDD127 pKa = 3.19AVCFNFDD134 pKa = 2.93RR135 pKa = 11.84WDD137 pKa = 3.6GFTGIAASEE146 pKa = 4.2SMSYY150 pKa = 10.97YY151 pKa = 8.36WGKK154 pKa = 10.82VNGGYY159 pKa = 10.35EE160 pKa = 4.2GAGKK164 pKa = 10.03YY165 pKa = 9.85ASCEE169 pKa = 4.07APDD172 pKa = 4.39DD173 pKa = 3.76ATAVITLSSPLPEE186 pKa = 4.01LVAALSLPAFSMQSPTALAEE206 pKa = 4.01YY207 pKa = 10.27GADD210 pKa = 3.12EE211 pKa = 4.51VGGSEE216 pKa = 5.19DD217 pKa = 4.66APTLPEE223 pKa = 4.16YY224 pKa = 10.39ATDD227 pKa = 3.97HH228 pKa = 6.1PTGTGPFTFEE238 pKa = 3.69SWSPGEE244 pKa = 4.15NVTLAANTEE253 pKa = 4.13YY254 pKa = 9.94WGEE257 pKa = 3.96QGQITTVIFPVISDD271 pKa = 3.56ATARR275 pKa = 11.84RR276 pKa = 11.84QALEE280 pKa = 4.12AGDD283 pKa = 3.54IDD285 pKa = 5.63GYY287 pKa = 11.33DD288 pKa = 3.43LVGPADD294 pKa = 3.76VVALEE299 pKa = 4.21DD300 pKa = 3.2AGYY303 pKa = 8.83TIEE306 pKa = 5.42NRR308 pKa = 11.84DD309 pKa = 3.96PFNVLYY315 pKa = 10.85LGMNQAVPEE324 pKa = 4.19LADD327 pKa = 3.07IRR329 pKa = 11.84VRR331 pKa = 11.84QAIAHH336 pKa = 6.96AIDD339 pKa = 3.84KK340 pKa = 9.98DD341 pKa = 3.83ALVAATLPEE350 pKa = 4.32GTLVATNFVPPSVRR364 pKa = 11.84GHH366 pKa = 6.2SDD368 pKa = 3.39DD369 pKa = 3.83VPTYY373 pKa = 11.21DD374 pKa = 4.58FDD376 pKa = 3.95QDD378 pKa = 3.37EE379 pKa = 4.61ARR381 pKa = 11.84SLLAEE386 pKa = 4.75AGVSDD391 pKa = 3.91LTLNFTYY398 pKa = 7.79PTNVSRR404 pKa = 11.84PYY406 pKa = 9.99MPTPEE411 pKa = 4.04QVFEE415 pKa = 4.97RR416 pKa = 11.84IAADD420 pKa = 3.64LEE422 pKa = 4.56EE423 pKa = 4.36VGITVEE429 pKa = 4.16ATPLPWSPDD438 pKa = 3.31YY439 pKa = 11.15LDD441 pKa = 5.65RR442 pKa = 11.84IQGGADD448 pKa = 3.12HH449 pKa = 7.02GLHH452 pKa = 6.13LLGWTGDD459 pKa = 3.72YY460 pKa = 11.07NDD462 pKa = 4.18TYY464 pKa = 11.73NFIGVFFGAQSNEE477 pKa = 3.57WGFDD481 pKa = 3.45DD482 pKa = 4.47PALFEE487 pKa = 4.47SLSTARR493 pKa = 11.84VATDD497 pKa = 3.22EE498 pKa = 4.59AEE500 pKa = 4.11QTSLYY505 pKa = 10.79EE506 pKa = 3.99EE507 pKa = 4.8ANVQIMEE514 pKa = 4.55NLPGIPIAHH523 pKa = 7.04PVPSLAFRR531 pKa = 11.84PGVEE535 pKa = 4.66NYY537 pKa = 7.8PASPVQDD544 pKa = 3.03EE545 pKa = 5.16VYY547 pKa = 11.09NVITLTDD554 pKa = 3.21
MM1 pKa = 7.39AVVGLAASSLVLAGCSSEE19 pKa = 4.22RR20 pKa = 11.84GEE22 pKa = 4.49GGSDD26 pKa = 3.2GSSSEE31 pKa = 4.25GGSGTSSEE39 pKa = 4.35TGEE42 pKa = 4.16AAGPTTFTFASSSDD56 pKa = 3.77PASLDD61 pKa = 3.26PAFASDD67 pKa = 3.92GEE69 pKa = 4.5SFRR72 pKa = 11.84VARR75 pKa = 11.84QMFEE79 pKa = 3.81GLVGVEE85 pKa = 4.84PGTADD90 pKa = 3.86PAPLLAEE97 pKa = 4.14SWDD100 pKa = 3.68VSDD103 pKa = 6.37DD104 pKa = 3.4GLEE107 pKa = 3.93YY108 pKa = 10.56TFQLKK113 pKa = 10.46EE114 pKa = 4.0GVSFHH119 pKa = 7.64DD120 pKa = 3.56GTPFNGDD127 pKa = 3.19AVCFNFDD134 pKa = 2.93RR135 pKa = 11.84WDD137 pKa = 3.6GFTGIAASEE146 pKa = 4.2SMSYY150 pKa = 10.97YY151 pKa = 8.36WGKK154 pKa = 10.82VNGGYY159 pKa = 10.35EE160 pKa = 4.2GAGKK164 pKa = 10.03YY165 pKa = 9.85ASCEE169 pKa = 4.07APDD172 pKa = 4.39DD173 pKa = 3.76ATAVITLSSPLPEE186 pKa = 4.01LVAALSLPAFSMQSPTALAEE206 pKa = 4.01YY207 pKa = 10.27GADD210 pKa = 3.12EE211 pKa = 4.51VGGSEE216 pKa = 5.19DD217 pKa = 4.66APTLPEE223 pKa = 4.16YY224 pKa = 10.39ATDD227 pKa = 3.97HH228 pKa = 6.1PTGTGPFTFEE238 pKa = 3.69SWSPGEE244 pKa = 4.15NVTLAANTEE253 pKa = 4.13YY254 pKa = 9.94WGEE257 pKa = 3.96QGQITTVIFPVISDD271 pKa = 3.56ATARR275 pKa = 11.84RR276 pKa = 11.84QALEE280 pKa = 4.12AGDD283 pKa = 3.54IDD285 pKa = 5.63GYY287 pKa = 11.33DD288 pKa = 3.43LVGPADD294 pKa = 3.76VVALEE299 pKa = 4.21DD300 pKa = 3.2AGYY303 pKa = 8.83TIEE306 pKa = 5.42NRR308 pKa = 11.84DD309 pKa = 3.96PFNVLYY315 pKa = 10.85LGMNQAVPEE324 pKa = 4.19LADD327 pKa = 3.07IRR329 pKa = 11.84VRR331 pKa = 11.84QAIAHH336 pKa = 6.96AIDD339 pKa = 3.84KK340 pKa = 9.98DD341 pKa = 3.83ALVAATLPEE350 pKa = 4.32GTLVATNFVPPSVRR364 pKa = 11.84GHH366 pKa = 6.2SDD368 pKa = 3.39DD369 pKa = 3.83VPTYY373 pKa = 11.21DD374 pKa = 4.58FDD376 pKa = 3.95QDD378 pKa = 3.37EE379 pKa = 4.61ARR381 pKa = 11.84SLLAEE386 pKa = 4.75AGVSDD391 pKa = 3.91LTLNFTYY398 pKa = 7.79PTNVSRR404 pKa = 11.84PYY406 pKa = 9.99MPTPEE411 pKa = 4.04QVFEE415 pKa = 4.97RR416 pKa = 11.84IAADD420 pKa = 3.64LEE422 pKa = 4.56EE423 pKa = 4.36VGITVEE429 pKa = 4.16ATPLPWSPDD438 pKa = 3.31YY439 pKa = 11.15LDD441 pKa = 5.65RR442 pKa = 11.84IQGGADD448 pKa = 3.12HH449 pKa = 7.02GLHH452 pKa = 6.13LLGWTGDD459 pKa = 3.72YY460 pKa = 11.07NDD462 pKa = 4.18TYY464 pKa = 11.73NFIGVFFGAQSNEE477 pKa = 3.57WGFDD481 pKa = 3.45DD482 pKa = 4.47PALFEE487 pKa = 4.47SLSTARR493 pKa = 11.84VATDD497 pKa = 3.22EE498 pKa = 4.59AEE500 pKa = 4.11QTSLYY505 pKa = 10.79EE506 pKa = 3.99EE507 pKa = 4.8ANVQIMEE514 pKa = 4.55NLPGIPIAHH523 pKa = 7.04PVPSLAFRR531 pKa = 11.84PGVEE535 pKa = 4.66NYY537 pKa = 7.8PASPVQDD544 pKa = 3.03EE545 pKa = 5.16VYY547 pKa = 11.09NVITLTDD554 pKa = 3.21
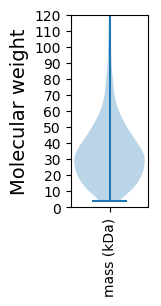
Molecular weight: 58.69 kDa
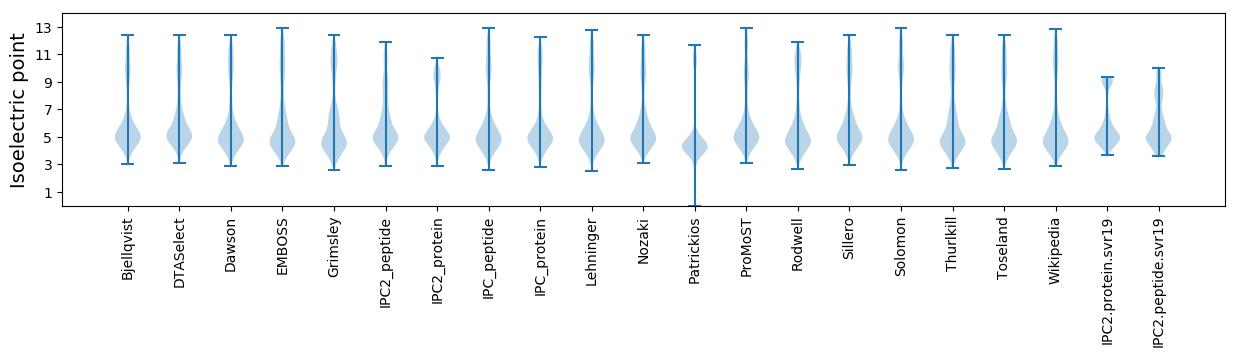
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Z1E3U8|A0A4Z1E3U8_9MICO Putative lipoprotein OS=Serinibacter arcticus OX=1655435 GN=SERN_1446 PE=4 SV=1
MM1 pKa = 7.56AVPKK5 pKa = 10.48RR6 pKa = 11.84KK7 pKa = 9.07MSRR10 pKa = 11.84SNTRR14 pKa = 11.84ARR16 pKa = 11.84RR17 pKa = 11.84SQWKK21 pKa = 8.13ATPATLSTCSKK32 pKa = 10.21CKK34 pKa = 10.33APRR37 pKa = 11.84LSHH40 pKa = 5.71QACPSCGAYY49 pKa = 9.97GEE51 pKa = 4.24RR52 pKa = 11.84TYY54 pKa = 11.79AEE56 pKa = 5.25AIRR59 pKa = 11.84SDD61 pKa = 3.24AA62 pKa = 4.14
MM1 pKa = 7.56AVPKK5 pKa = 10.48RR6 pKa = 11.84KK7 pKa = 9.07MSRR10 pKa = 11.84SNTRR14 pKa = 11.84ARR16 pKa = 11.84RR17 pKa = 11.84SQWKK21 pKa = 8.13ATPATLSTCSKK32 pKa = 10.21CKK34 pKa = 10.33APRR37 pKa = 11.84LSHH40 pKa = 5.71QACPSCGAYY49 pKa = 9.97GEE51 pKa = 4.24RR52 pKa = 11.84TYY54 pKa = 11.79AEE56 pKa = 5.25AIRR59 pKa = 11.84SDD61 pKa = 3.24AA62 pKa = 4.14
Molecular weight: 6.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1041505 |
37 |
3780 |
347.3 |
36.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.299 ± 0.062 | 0.484 ± 0.009 |
6.436 ± 0.038 | 5.484 ± 0.039 |
2.547 ± 0.029 | 9.567 ± 0.042 |
1.987 ± 0.025 | 3.366 ± 0.031 |
1.253 ± 0.023 | 10.172 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.568 ± 0.016 | 1.582 ± 0.028 |
5.892 ± 0.042 | 2.562 ± 0.021 |
7.518 ± 0.061 | 5.418 ± 0.03 |
6.698 ± 0.058 | 9.926 ± 0.049 |
1.48 ± 0.019 | 1.764 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
