
Ehrlichia chaffeensis (strain ATCC CRL-10679 / Arkansas)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rickettsiales; Anaplasmataceae; Ehrlichia; canis group; Ehrlichia chaffeensis
Average proteome isoelectric point is 6.95
Get precalculated fractions of proteins
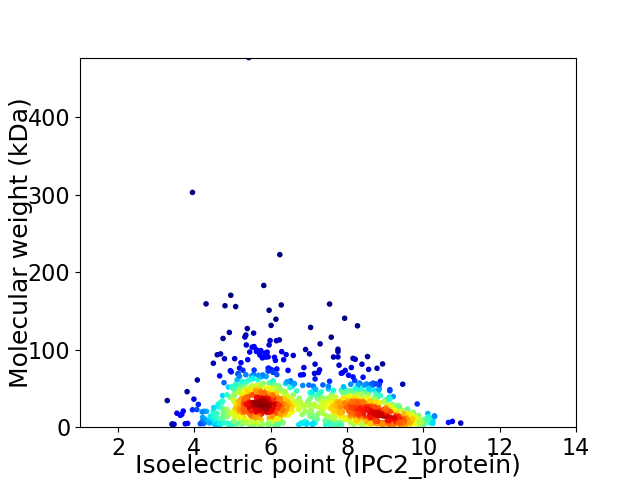
Virtual 2D-PAGE plot for 1100 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q2GFX2|Q2GFX2_EHRCR Uncharacterized protein OS=Ehrlichia chaffeensis (strain ATCC CRL-10679 / Arkansas) OX=205920 GN=ECH_0866 PE=4 SV=1
MM1 pKa = 7.46NNTHH5 pKa = 5.63NTNNVVNQVQDD16 pKa = 3.24IAQDD20 pKa = 3.43NHH22 pKa = 7.96ISDD25 pKa = 4.08NNANEE30 pKa = 3.82ARR32 pKa = 11.84EE33 pKa = 3.99VLAAHH38 pKa = 6.9AAEE41 pKa = 4.39NNANEE46 pKa = 4.12IRR48 pKa = 11.84EE49 pKa = 4.12VLHH52 pKa = 7.43DD53 pKa = 4.19YY54 pKa = 11.37SLDD57 pKa = 3.7DD58 pKa = 4.58YY59 pKa = 11.72AIDD62 pKa = 3.69DD63 pKa = 4.27HH64 pKa = 7.86NIGGNAEE71 pKa = 4.04AQDD74 pKa = 4.7DD75 pKa = 4.41YY76 pKa = 12.12SLDD79 pKa = 3.91DD80 pKa = 4.08YY81 pKa = 11.81AIDD84 pKa = 5.06DD85 pKa = 4.07YY86 pKa = 11.87NIGGNAEE93 pKa = 4.05VQDD96 pKa = 3.78NHH98 pKa = 8.18ISDD101 pKa = 4.1NNTNEE106 pKa = 3.65ARR108 pKa = 11.84EE109 pKa = 4.02VLLAHH114 pKa = 7.02AAEE117 pKa = 4.42NNANEE122 pKa = 4.11IRR124 pKa = 11.84EE125 pKa = 4.02VLYY128 pKa = 10.54DD129 pKa = 3.76YY130 pKa = 11.6SLDD133 pKa = 4.07DD134 pKa = 4.54YY135 pKa = 11.68AIDD138 pKa = 4.31DD139 pKa = 4.31HH140 pKa = 8.24NIDD143 pKa = 4.16GNAEE147 pKa = 3.96VQDD150 pKa = 3.68NHH152 pKa = 8.18ISDD155 pKa = 4.1NNTNEE160 pKa = 3.65ARR162 pKa = 11.84EE163 pKa = 4.02VLLAHH168 pKa = 7.02AAEE171 pKa = 4.42NNANEE176 pKa = 4.11IRR178 pKa = 11.84EE179 pKa = 4.02VLYY182 pKa = 10.54DD183 pKa = 3.76YY184 pKa = 11.6SLDD187 pKa = 4.07DD188 pKa = 4.54YY189 pKa = 11.68AIDD192 pKa = 3.69DD193 pKa = 4.27HH194 pKa = 7.86NIGGNAEE201 pKa = 4.04AQDD204 pKa = 4.7DD205 pKa = 4.41YY206 pKa = 12.12SLDD209 pKa = 3.91DD210 pKa = 4.08YY211 pKa = 11.81AIDD214 pKa = 5.06DD215 pKa = 4.07YY216 pKa = 11.87NIGGNAEE223 pKa = 3.99AQDD226 pKa = 4.11NYY228 pKa = 11.59SLDD231 pKa = 4.13DD232 pKa = 3.91YY233 pKa = 11.73AIDD236 pKa = 5.06DD237 pKa = 4.07YY238 pKa = 11.87NIGGNAEE245 pKa = 4.05VQDD248 pKa = 3.78NHH250 pKa = 8.18ISDD253 pKa = 4.0NNTNEE258 pKa = 3.8TRR260 pKa = 11.84EE261 pKa = 4.1ALAAHH266 pKa = 6.58AAEE269 pKa = 4.45NNANEE274 pKa = 4.05IRR276 pKa = 11.84EE277 pKa = 4.01VLYY280 pKa = 10.83NYY282 pKa = 10.84SLDD285 pKa = 4.41DD286 pKa = 4.1YY287 pKa = 11.56AIDD290 pKa = 4.31DD291 pKa = 4.31HH292 pKa = 8.24NIDD295 pKa = 4.16GNAEE299 pKa = 3.96VQDD302 pKa = 3.68NHH304 pKa = 8.18ISDD307 pKa = 4.0NNTNEE312 pKa = 3.8TRR314 pKa = 11.84EE315 pKa = 4.1ALAAHH320 pKa = 6.58AAEE323 pKa = 4.45NNANEE328 pKa = 4.05IRR330 pKa = 11.84EE331 pKa = 4.01VLYY334 pKa = 10.83NYY336 pKa = 10.84SLDD339 pKa = 4.41DD340 pKa = 4.1YY341 pKa = 11.56AIDD344 pKa = 4.31DD345 pKa = 4.31HH346 pKa = 8.24NIDD349 pKa = 4.16GNAEE353 pKa = 3.96VQDD356 pKa = 3.68NHH358 pKa = 8.18ISDD361 pKa = 4.1NNTNEE366 pKa = 3.66ARR368 pKa = 11.84EE369 pKa = 4.09AQVNEE374 pKa = 3.98EE375 pKa = 4.29LDD377 pKa = 3.99NVNTTANTQNINEE390 pKa = 4.34RR391 pKa = 11.84LEE393 pKa = 4.31HH394 pKa = 5.71CHH396 pKa = 6.68ISQTNLHH403 pKa = 6.11KK404 pKa = 10.17HH405 pKa = 5.4TFQKK409 pKa = 11.07
MM1 pKa = 7.46NNTHH5 pKa = 5.63NTNNVVNQVQDD16 pKa = 3.24IAQDD20 pKa = 3.43NHH22 pKa = 7.96ISDD25 pKa = 4.08NNANEE30 pKa = 3.82ARR32 pKa = 11.84EE33 pKa = 3.99VLAAHH38 pKa = 6.9AAEE41 pKa = 4.39NNANEE46 pKa = 4.12IRR48 pKa = 11.84EE49 pKa = 4.12VLHH52 pKa = 7.43DD53 pKa = 4.19YY54 pKa = 11.37SLDD57 pKa = 3.7DD58 pKa = 4.58YY59 pKa = 11.72AIDD62 pKa = 3.69DD63 pKa = 4.27HH64 pKa = 7.86NIGGNAEE71 pKa = 4.04AQDD74 pKa = 4.7DD75 pKa = 4.41YY76 pKa = 12.12SLDD79 pKa = 3.91DD80 pKa = 4.08YY81 pKa = 11.81AIDD84 pKa = 5.06DD85 pKa = 4.07YY86 pKa = 11.87NIGGNAEE93 pKa = 4.05VQDD96 pKa = 3.78NHH98 pKa = 8.18ISDD101 pKa = 4.1NNTNEE106 pKa = 3.65ARR108 pKa = 11.84EE109 pKa = 4.02VLLAHH114 pKa = 7.02AAEE117 pKa = 4.42NNANEE122 pKa = 4.11IRR124 pKa = 11.84EE125 pKa = 4.02VLYY128 pKa = 10.54DD129 pKa = 3.76YY130 pKa = 11.6SLDD133 pKa = 4.07DD134 pKa = 4.54YY135 pKa = 11.68AIDD138 pKa = 4.31DD139 pKa = 4.31HH140 pKa = 8.24NIDD143 pKa = 4.16GNAEE147 pKa = 3.96VQDD150 pKa = 3.68NHH152 pKa = 8.18ISDD155 pKa = 4.1NNTNEE160 pKa = 3.65ARR162 pKa = 11.84EE163 pKa = 4.02VLLAHH168 pKa = 7.02AAEE171 pKa = 4.42NNANEE176 pKa = 4.11IRR178 pKa = 11.84EE179 pKa = 4.02VLYY182 pKa = 10.54DD183 pKa = 3.76YY184 pKa = 11.6SLDD187 pKa = 4.07DD188 pKa = 4.54YY189 pKa = 11.68AIDD192 pKa = 3.69DD193 pKa = 4.27HH194 pKa = 7.86NIGGNAEE201 pKa = 4.04AQDD204 pKa = 4.7DD205 pKa = 4.41YY206 pKa = 12.12SLDD209 pKa = 3.91DD210 pKa = 4.08YY211 pKa = 11.81AIDD214 pKa = 5.06DD215 pKa = 4.07YY216 pKa = 11.87NIGGNAEE223 pKa = 3.99AQDD226 pKa = 4.11NYY228 pKa = 11.59SLDD231 pKa = 4.13DD232 pKa = 3.91YY233 pKa = 11.73AIDD236 pKa = 5.06DD237 pKa = 4.07YY238 pKa = 11.87NIGGNAEE245 pKa = 4.05VQDD248 pKa = 3.78NHH250 pKa = 8.18ISDD253 pKa = 4.0NNTNEE258 pKa = 3.8TRR260 pKa = 11.84EE261 pKa = 4.1ALAAHH266 pKa = 6.58AAEE269 pKa = 4.45NNANEE274 pKa = 4.05IRR276 pKa = 11.84EE277 pKa = 4.01VLYY280 pKa = 10.83NYY282 pKa = 10.84SLDD285 pKa = 4.41DD286 pKa = 4.1YY287 pKa = 11.56AIDD290 pKa = 4.31DD291 pKa = 4.31HH292 pKa = 8.24NIDD295 pKa = 4.16GNAEE299 pKa = 3.96VQDD302 pKa = 3.68NHH304 pKa = 8.18ISDD307 pKa = 4.0NNTNEE312 pKa = 3.8TRR314 pKa = 11.84EE315 pKa = 4.1ALAAHH320 pKa = 6.58AAEE323 pKa = 4.45NNANEE328 pKa = 4.05IRR330 pKa = 11.84EE331 pKa = 4.01VLYY334 pKa = 10.83NYY336 pKa = 10.84SLDD339 pKa = 4.41DD340 pKa = 4.1YY341 pKa = 11.56AIDD344 pKa = 4.31DD345 pKa = 4.31HH346 pKa = 8.24NIDD349 pKa = 4.16GNAEE353 pKa = 3.96VQDD356 pKa = 3.68NHH358 pKa = 8.18ISDD361 pKa = 4.1NNTNEE366 pKa = 3.66ARR368 pKa = 11.84EE369 pKa = 4.09AQVNEE374 pKa = 3.98EE375 pKa = 4.29LDD377 pKa = 3.99NVNTTANTQNINEE390 pKa = 4.34RR391 pKa = 11.84LEE393 pKa = 4.31HH394 pKa = 5.71CHH396 pKa = 6.68ISQTNLHH403 pKa = 6.11KK404 pKa = 10.17HH405 pKa = 5.4TFQKK409 pKa = 11.07
Molecular weight: 45.86 kDa
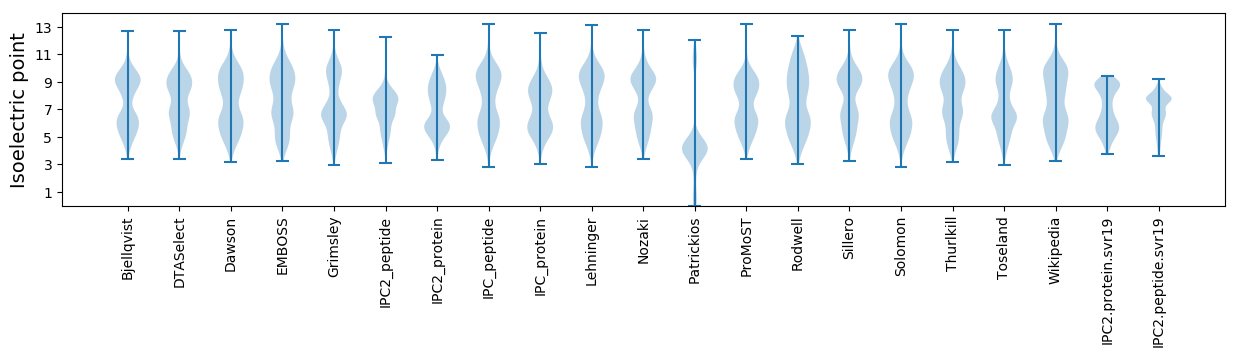
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q2GH34|RS11_EHRCR 30S ribosomal protein S11 OS=Ehrlichia chaffeensis (strain ATCC CRL-10679 / Arkansas) OX=205920 GN=rpsK PE=3 SV=1
MM1 pKa = 7.71KK2 pKa = 9.6GTFQPSRR9 pKa = 11.84IVRR12 pKa = 11.84KK13 pKa = 9.02RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.86GFRR19 pKa = 11.84SRR21 pKa = 11.84MSTKK25 pKa = 9.02MGRR28 pKa = 11.84RR29 pKa = 11.84ILNRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84RR40 pKa = 11.84VLCAA44 pKa = 3.56
MM1 pKa = 7.71KK2 pKa = 9.6GTFQPSRR9 pKa = 11.84IVRR12 pKa = 11.84KK13 pKa = 9.02RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.86GFRR19 pKa = 11.84SRR21 pKa = 11.84MSTKK25 pKa = 9.02MGRR28 pKa = 11.84RR29 pKa = 11.84ILNRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84RR40 pKa = 11.84VLCAA44 pKa = 3.56
Molecular weight: 5.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
310362 |
30 |
4313 |
282.1 |
31.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.37 ± 0.067 | 1.669 ± 0.04 |
5.434 ± 0.101 | 5.495 ± 0.087 |
4.541 ± 0.075 | 5.501 ± 0.067 |
2.224 ± 0.036 | 9.747 ± 0.131 |
7.233 ± 0.069 | 9.551 ± 0.086 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.457 ± 0.039 | 6.499 ± 0.091 |
3.021 ± 0.043 | 3.327 ± 0.049 |
3.532 ± 0.052 | 8.039 ± 0.092 |
4.865 ± 0.064 | 6.876 ± 0.078 |
0.652 ± 0.023 | 3.965 ± 0.062 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
