
Vibrio phage JA-1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Schitoviridae; Pacinivirus; unclassified Pacinivirus
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
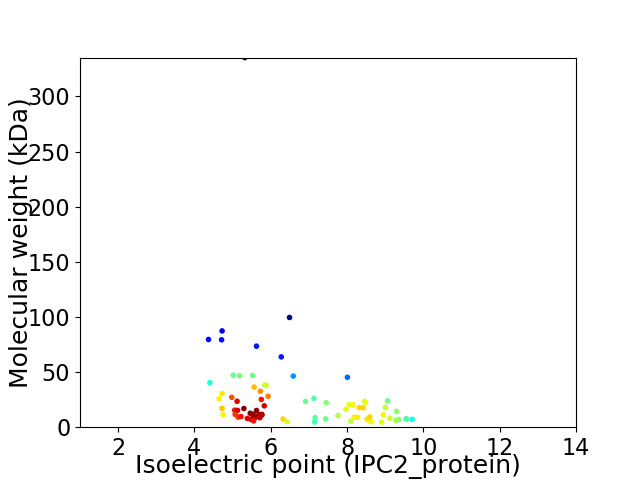
Virtual 2D-PAGE plot for 79 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R9R5B6|R9R5B6_9CAUD Uncharacterized protein OS=Vibrio phage JA-1 OX=1283071 GN=JA1_0061 PE=4 SV=1
MM1 pKa = 8.09DD2 pKa = 3.99NHH4 pKa = 6.95PFFIMGLFSKK14 pKa = 9.33TRR16 pKa = 11.84IYY18 pKa = 10.39TYY20 pKa = 10.43SQTVSLIEE28 pKa = 4.09DD29 pKa = 3.7TPDD32 pKa = 3.19VVKK35 pKa = 11.02QSVTNSILGNKK46 pKa = 9.21EE47 pKa = 3.52IVPNLMDD54 pKa = 3.32TTLNRR59 pKa = 11.84LSSRR63 pKa = 11.84VNRR66 pKa = 11.84YY67 pKa = 8.39YY68 pKa = 10.98KK69 pKa = 10.75YY70 pKa = 10.83GITDD74 pKa = 4.07FTNGLPEE81 pKa = 4.17GSFGYY86 pKa = 10.42FKK88 pKa = 11.14ADD90 pKa = 3.1ADD92 pKa = 3.91AVIDD96 pKa = 3.67VLTNHH101 pKa = 6.03VLTLQPNEE109 pKa = 4.1EE110 pKa = 4.21VSLISYY116 pKa = 9.09VIGEE120 pKa = 4.54INPAFYY126 pKa = 10.56GFKK129 pKa = 9.74WLRR132 pKa = 11.84GNTDD136 pKa = 2.33WNYY139 pKa = 10.32KK140 pKa = 10.08DD141 pKa = 4.8NYY143 pKa = 7.83VTTVPGIGTNNLVYY157 pKa = 10.8AGFDD161 pKa = 3.33VLSEE165 pKa = 4.01NLIKK169 pKa = 11.17LNFNEE174 pKa = 4.26QNDD177 pKa = 4.33LSPEE181 pKa = 4.04PNVYY185 pKa = 10.44SITIPAVPSITDD197 pKa = 3.18EE198 pKa = 4.2LYY200 pKa = 11.56YY201 pKa = 10.2MVTYY205 pKa = 10.88NIVDD209 pKa = 3.86KK210 pKa = 10.28DD211 pKa = 3.84TEE213 pKa = 4.15DD214 pKa = 5.03AIGNPYY220 pKa = 9.1YY221 pKa = 10.16WIYY224 pKa = 11.0NAAEE228 pKa = 4.63DD229 pKa = 4.34IYY231 pKa = 11.69VEE233 pKa = 4.33LKK235 pKa = 10.43LPEE238 pKa = 3.99YY239 pKa = 10.64SKK241 pKa = 9.94YY242 pKa = 8.0TQYY245 pKa = 11.62LPIVPVRR252 pKa = 11.84IQNQDD257 pKa = 3.13MLDD260 pKa = 3.88PNLTTDD266 pKa = 4.65LYY268 pKa = 8.87RR269 pKa = 11.84TSVKK273 pKa = 9.67ILKK276 pKa = 10.24YY277 pKa = 10.71VDD279 pKa = 3.01IDD281 pKa = 3.89ANQLRR286 pKa = 11.84EE287 pKa = 4.54SINASPDD294 pKa = 3.09IGSLDD299 pKa = 2.95HH300 pKa = 7.04AYY302 pKa = 10.28FVFGYY307 pKa = 8.23PIRR310 pKa = 11.84VEE312 pKa = 3.79SDD314 pKa = 2.86VGNEE318 pKa = 3.54YY319 pKa = 10.64LYY321 pKa = 10.74EE322 pKa = 4.09YY323 pKa = 10.85FNDD326 pKa = 3.24LHH328 pKa = 6.86DD329 pKa = 4.99RR330 pKa = 11.84NIFTKK335 pKa = 10.44TDD337 pKa = 3.46FDD339 pKa = 3.53NWFSSEE345 pKa = 4.35TEE347 pKa = 3.92KK348 pKa = 8.43TTPPVNVIQIEE359 pKa = 4.18DD360 pKa = 3.58DD361 pKa = 3.72TYY363 pKa = 9.75KK364 pKa = 10.54TKK366 pKa = 10.59IMYY369 pKa = 9.98YY370 pKa = 9.95YY371 pKa = 8.36ITRR374 pKa = 11.84EE375 pKa = 3.87IKK377 pKa = 10.41VGNLTKK383 pKa = 9.93IGKK386 pKa = 9.09VIKK389 pKa = 9.76EE390 pKa = 4.29VYY392 pKa = 10.16SGNQDD397 pKa = 2.56ITFEE401 pKa = 4.25NDD403 pKa = 2.64NVSYY407 pKa = 8.35TTEE410 pKa = 4.01SPDD413 pKa = 3.23NRR415 pKa = 11.84VVLKK419 pKa = 10.73KK420 pKa = 10.61QITTNTYY427 pKa = 9.79SEE429 pKa = 4.89LIITGLVHH437 pKa = 6.78FNNIYY442 pKa = 10.26GVNRR446 pKa = 11.84DD447 pKa = 4.2EE448 pKa = 4.54ITTLDD453 pKa = 3.65SANTEE458 pKa = 4.15SFIIPLNISIINQMKK473 pKa = 10.17LWDD476 pKa = 4.67RR477 pKa = 11.84DD478 pKa = 3.51DD479 pKa = 5.75LYY481 pKa = 11.62YY482 pKa = 11.03KK483 pKa = 10.62CFNLVLNAYY492 pKa = 8.95EE493 pKa = 4.31KK494 pKa = 10.63IKK496 pKa = 10.73LKK498 pKa = 9.99WYY500 pKa = 6.68QTGIFKK506 pKa = 10.73VFTIVLVIAISIYY519 pKa = 10.02TGGAASFITGLISAATAGVISLGIFIAEE547 pKa = 4.96LYY549 pKa = 7.6LTSYY553 pKa = 8.62VFKK556 pKa = 11.3YY557 pKa = 10.49SFEE560 pKa = 4.24KK561 pKa = 10.54VAEE564 pKa = 4.01ALGIDD569 pKa = 3.96LAFVLALVLVIGGSIASLNSLPNADD594 pKa = 3.02IFLNVGTGLGQGVGQVIQEE613 pKa = 4.1EE614 pKa = 4.57AIGIQDD620 pKa = 4.14EE621 pKa = 4.46LTALQEE627 pKa = 4.19YY628 pKa = 10.38YY629 pKa = 10.95GLKK632 pKa = 9.92FDD634 pKa = 4.83EE635 pKa = 4.87LFSVQEE641 pKa = 3.87EE642 pKa = 4.5LQTNKK647 pKa = 9.86TNILDD652 pKa = 4.67PIGLYY657 pKa = 9.93TDD659 pKa = 3.23IGMFPNEE666 pKa = 4.16SSEE669 pKa = 4.19MYY671 pKa = 9.92LQRR674 pKa = 11.84KK675 pKa = 8.67IEE677 pKa = 3.99LVNPGILTIDD687 pKa = 3.35AVSNFVDD694 pKa = 3.46NMLTLRR700 pKa = 11.84TSS702 pKa = 3.03
MM1 pKa = 8.09DD2 pKa = 3.99NHH4 pKa = 6.95PFFIMGLFSKK14 pKa = 9.33TRR16 pKa = 11.84IYY18 pKa = 10.39TYY20 pKa = 10.43SQTVSLIEE28 pKa = 4.09DD29 pKa = 3.7TPDD32 pKa = 3.19VVKK35 pKa = 11.02QSVTNSILGNKK46 pKa = 9.21EE47 pKa = 3.52IVPNLMDD54 pKa = 3.32TTLNRR59 pKa = 11.84LSSRR63 pKa = 11.84VNRR66 pKa = 11.84YY67 pKa = 8.39YY68 pKa = 10.98KK69 pKa = 10.75YY70 pKa = 10.83GITDD74 pKa = 4.07FTNGLPEE81 pKa = 4.17GSFGYY86 pKa = 10.42FKK88 pKa = 11.14ADD90 pKa = 3.1ADD92 pKa = 3.91AVIDD96 pKa = 3.67VLTNHH101 pKa = 6.03VLTLQPNEE109 pKa = 4.1EE110 pKa = 4.21VSLISYY116 pKa = 9.09VIGEE120 pKa = 4.54INPAFYY126 pKa = 10.56GFKK129 pKa = 9.74WLRR132 pKa = 11.84GNTDD136 pKa = 2.33WNYY139 pKa = 10.32KK140 pKa = 10.08DD141 pKa = 4.8NYY143 pKa = 7.83VTTVPGIGTNNLVYY157 pKa = 10.8AGFDD161 pKa = 3.33VLSEE165 pKa = 4.01NLIKK169 pKa = 11.17LNFNEE174 pKa = 4.26QNDD177 pKa = 4.33LSPEE181 pKa = 4.04PNVYY185 pKa = 10.44SITIPAVPSITDD197 pKa = 3.18EE198 pKa = 4.2LYY200 pKa = 11.56YY201 pKa = 10.2MVTYY205 pKa = 10.88NIVDD209 pKa = 3.86KK210 pKa = 10.28DD211 pKa = 3.84TEE213 pKa = 4.15DD214 pKa = 5.03AIGNPYY220 pKa = 9.1YY221 pKa = 10.16WIYY224 pKa = 11.0NAAEE228 pKa = 4.63DD229 pKa = 4.34IYY231 pKa = 11.69VEE233 pKa = 4.33LKK235 pKa = 10.43LPEE238 pKa = 3.99YY239 pKa = 10.64SKK241 pKa = 9.94YY242 pKa = 8.0TQYY245 pKa = 11.62LPIVPVRR252 pKa = 11.84IQNQDD257 pKa = 3.13MLDD260 pKa = 3.88PNLTTDD266 pKa = 4.65LYY268 pKa = 8.87RR269 pKa = 11.84TSVKK273 pKa = 9.67ILKK276 pKa = 10.24YY277 pKa = 10.71VDD279 pKa = 3.01IDD281 pKa = 3.89ANQLRR286 pKa = 11.84EE287 pKa = 4.54SINASPDD294 pKa = 3.09IGSLDD299 pKa = 2.95HH300 pKa = 7.04AYY302 pKa = 10.28FVFGYY307 pKa = 8.23PIRR310 pKa = 11.84VEE312 pKa = 3.79SDD314 pKa = 2.86VGNEE318 pKa = 3.54YY319 pKa = 10.64LYY321 pKa = 10.74EE322 pKa = 4.09YY323 pKa = 10.85FNDD326 pKa = 3.24LHH328 pKa = 6.86DD329 pKa = 4.99RR330 pKa = 11.84NIFTKK335 pKa = 10.44TDD337 pKa = 3.46FDD339 pKa = 3.53NWFSSEE345 pKa = 4.35TEE347 pKa = 3.92KK348 pKa = 8.43TTPPVNVIQIEE359 pKa = 4.18DD360 pKa = 3.58DD361 pKa = 3.72TYY363 pKa = 9.75KK364 pKa = 10.54TKK366 pKa = 10.59IMYY369 pKa = 9.98YY370 pKa = 9.95YY371 pKa = 8.36ITRR374 pKa = 11.84EE375 pKa = 3.87IKK377 pKa = 10.41VGNLTKK383 pKa = 9.93IGKK386 pKa = 9.09VIKK389 pKa = 9.76EE390 pKa = 4.29VYY392 pKa = 10.16SGNQDD397 pKa = 2.56ITFEE401 pKa = 4.25NDD403 pKa = 2.64NVSYY407 pKa = 8.35TTEE410 pKa = 4.01SPDD413 pKa = 3.23NRR415 pKa = 11.84VVLKK419 pKa = 10.73KK420 pKa = 10.61QITTNTYY427 pKa = 9.79SEE429 pKa = 4.89LIITGLVHH437 pKa = 6.78FNNIYY442 pKa = 10.26GVNRR446 pKa = 11.84DD447 pKa = 4.2EE448 pKa = 4.54ITTLDD453 pKa = 3.65SANTEE458 pKa = 4.15SFIIPLNISIINQMKK473 pKa = 10.17LWDD476 pKa = 4.67RR477 pKa = 11.84DD478 pKa = 3.51DD479 pKa = 5.75LYY481 pKa = 11.62YY482 pKa = 11.03KK483 pKa = 10.62CFNLVLNAYY492 pKa = 8.95EE493 pKa = 4.31KK494 pKa = 10.63IKK496 pKa = 10.73LKK498 pKa = 9.99WYY500 pKa = 6.68QTGIFKK506 pKa = 10.73VFTIVLVIAISIYY519 pKa = 10.02TGGAASFITGLISAATAGVISLGIFIAEE547 pKa = 4.96LYY549 pKa = 7.6LTSYY553 pKa = 8.62VFKK556 pKa = 11.3YY557 pKa = 10.49SFEE560 pKa = 4.24KK561 pKa = 10.54VAEE564 pKa = 4.01ALGIDD569 pKa = 3.96LAFVLALVLVIGGSIASLNSLPNADD594 pKa = 3.02IFLNVGTGLGQGVGQVIQEE613 pKa = 4.1EE614 pKa = 4.57AIGIQDD620 pKa = 4.14EE621 pKa = 4.46LTALQEE627 pKa = 4.19YY628 pKa = 10.38YY629 pKa = 10.95GLKK632 pKa = 9.92FDD634 pKa = 4.83EE635 pKa = 4.87LFSVQEE641 pKa = 3.87EE642 pKa = 4.5LQTNKK647 pKa = 9.86TNILDD652 pKa = 4.67PIGLYY657 pKa = 9.93TDD659 pKa = 3.23IGMFPNEE666 pKa = 4.16SSEE669 pKa = 4.19MYY671 pKa = 9.92LQRR674 pKa = 11.84KK675 pKa = 8.67IEE677 pKa = 3.99LVNPGILTIDD687 pKa = 3.35AVSNFVDD694 pKa = 3.46NMLTLRR700 pKa = 11.84TSS702 pKa = 3.03
Molecular weight: 79.57 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R9R4F7|R9R4F7_9CAUD PF10947 family protein OS=Vibrio phage JA-1 OX=1283071 GN=JA1_0018 PE=4 SV=1
MM1 pKa = 7.64KK2 pKa = 10.5LLNVWVHH9 pKa = 4.71PTEE12 pKa = 4.46PEE14 pKa = 3.75SWFHH18 pKa = 5.35YY19 pKa = 9.88FKK21 pKa = 11.34VNNQVIRR28 pKa = 11.84FIPSPKK34 pKa = 9.75HH35 pKa = 6.38RR36 pKa = 11.84INNCPLRR43 pKa = 11.84IIPNHH48 pKa = 5.56FVKK51 pKa = 10.53LDD53 pKa = 3.43KK54 pKa = 10.92QLTVLRR60 pKa = 11.84TVV62 pKa = 3.24
MM1 pKa = 7.64KK2 pKa = 10.5LLNVWVHH9 pKa = 4.71PTEE12 pKa = 4.46PEE14 pKa = 3.75SWFHH18 pKa = 5.35YY19 pKa = 9.88FKK21 pKa = 11.34VNNQVIRR28 pKa = 11.84FIPSPKK34 pKa = 9.75HH35 pKa = 6.38RR36 pKa = 11.84INNCPLRR43 pKa = 11.84IIPNHH48 pKa = 5.56FVKK51 pKa = 10.53LDD53 pKa = 3.43KK54 pKa = 10.92QLTVLRR60 pKa = 11.84TVV62 pKa = 3.24
Molecular weight: 7.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
18339 |
37 |
3022 |
232.1 |
26.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.587 ± 0.428 | 0.878 ± 0.174 |
5.595 ± 0.2 | 7.307 ± 0.34 |
4.03 ± 0.167 | 5.595 ± 0.289 |
1.865 ± 0.232 | 6.587 ± 0.313 |
7.879 ± 0.273 | 8.91 ± 0.228 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.585 ± 0.179 | 6.304 ± 0.178 |
3.659 ± 0.162 | 4.051 ± 0.274 |
3.79 ± 0.167 | 6.205 ± 0.283 |
6.429 ± 0.173 | 6.347 ± 0.284 |
1.14 ± 0.159 | 4.259 ± 0.282 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
