
Microbacterium esteraromaticum
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Microbacterium
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
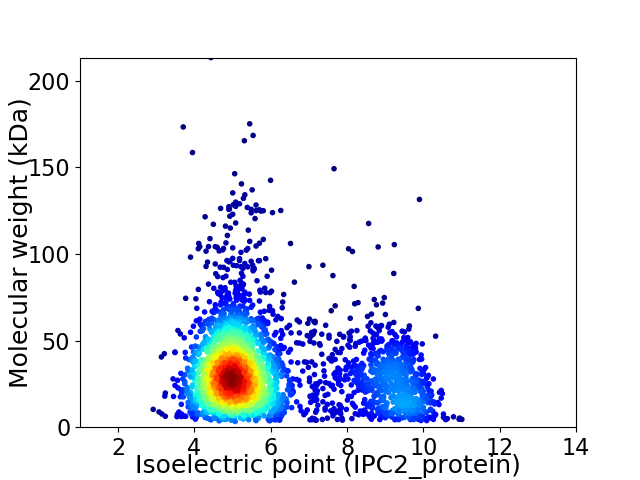
Virtual 2D-PAGE plot for 3202 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1R4JY53|A0A1R4JY53_9MICO Uncharacterized protein OS=Microbacterium esteraromaticum OX=57043 GN=FM104_09385 PE=4 SV=1
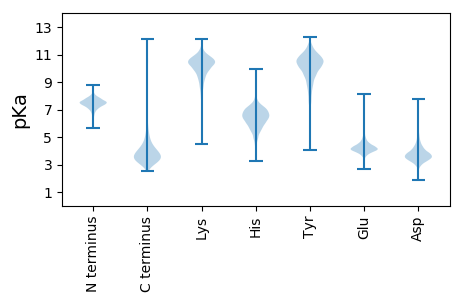
MM1 pKa = 7.56RR2 pKa = 11.84AVKK5 pKa = 9.33RR6 pKa = 11.84TGLAVLASAALGLGALVATPAYY28 pKa = 9.8AATVDD33 pKa = 4.06DD34 pKa = 5.13VIDD37 pKa = 4.81GIIYY41 pKa = 9.89EE42 pKa = 4.68ADD44 pKa = 3.77DD45 pKa = 4.36ADD47 pKa = 4.03VAAGATVTGFDD58 pKa = 3.62GTLTTVNIPDD68 pKa = 3.62TVAIEE73 pKa = 4.02GTEE76 pKa = 3.96YY77 pKa = 11.21DD78 pKa = 3.8VVAIGNSAFVTYY90 pKa = 10.35RR91 pKa = 11.84SGGVITQGDD100 pKa = 3.55ITEE103 pKa = 4.28VVIGDD108 pKa = 3.61NVRR111 pKa = 11.84TIGLQAFMSNRR122 pKa = 11.84IDD124 pKa = 3.74SLTLGSSVTSIKK136 pKa = 10.66GSAFMGNSLTHH147 pKa = 5.25ITIPSAVTSIGGAAFAEE164 pKa = 4.15NDD166 pKa = 3.76DD167 pKa = 4.03LVSVVFTGPAPTVRR181 pKa = 11.84EE182 pKa = 3.7ADD184 pKa = 3.18AGYY187 pKa = 10.76SSASFSTRR195 pKa = 11.84DD196 pKa = 3.27TEE198 pKa = 4.28FVLRR202 pKa = 11.84FPEE205 pKa = 4.5QYY207 pKa = 10.62SADD210 pKa = 3.8NVAGGYY216 pKa = 6.55TTPLWFGYY224 pKa = 5.19TTEE227 pKa = 5.52AIGDD231 pKa = 4.09DD232 pKa = 4.18EE233 pKa = 5.76PDD235 pKa = 3.28ASLSITKK242 pKa = 8.87TVDD245 pKa = 2.68ASFVRR250 pKa = 11.84VYY252 pKa = 11.04DD253 pKa = 3.25WDD255 pKa = 3.9FEE257 pKa = 4.47KK258 pKa = 10.39TGTVGAITVDD268 pKa = 3.41GDD270 pKa = 3.53EE271 pKa = 4.81ASADD275 pKa = 3.24VDD277 pKa = 4.17FTVTATPNGYY287 pKa = 9.43VDD289 pKa = 3.43SYY291 pKa = 11.39YY292 pKa = 11.17VLEE295 pKa = 4.26GTITVLNEE303 pKa = 3.34GTEE306 pKa = 4.11AQTVDD311 pKa = 3.75VVDD314 pKa = 4.37VPDD317 pKa = 3.7VAASASCSVYY327 pKa = 10.74DD328 pKa = 5.1SSFQPAEE335 pKa = 3.99GVEE338 pKa = 4.25IAASEE343 pKa = 4.23SAEE346 pKa = 4.01FEE348 pKa = 4.26YY349 pKa = 10.85SCYY352 pKa = 10.22FDD354 pKa = 3.79GTPEE358 pKa = 4.99DD359 pKa = 4.02GTNTATVTWGDD370 pKa = 3.9DD371 pKa = 3.41QSASATADD379 pKa = 3.31VVFTQRR385 pKa = 11.84DD386 pKa = 3.49TSFQTITIGDD396 pKa = 3.77DD397 pKa = 3.25HH398 pKa = 7.73ADD400 pKa = 3.27PDD402 pKa = 4.6GPFEE406 pKa = 4.12EE407 pKa = 5.75LGTAEE412 pKa = 4.55WNSDD416 pKa = 3.36HH417 pKa = 7.1LPIEE421 pKa = 4.14FTYY424 pKa = 10.86TLTADD429 pKa = 3.49GLTVGEE435 pKa = 4.86CTTLTNTAAIYY446 pKa = 10.05EE447 pKa = 4.34MEE449 pKa = 4.61RR450 pKa = 11.84ATQASVEE457 pKa = 4.24ICPEE461 pKa = 3.92PAVVIPDD468 pKa = 4.33PEE470 pKa = 4.23PTPDD474 pKa = 4.42PEE476 pKa = 4.25PTPDD480 pKa = 4.55PEE482 pKa = 4.47PSPAPVVDD490 pKa = 4.56PEE492 pKa = 4.22PAGLATTGTDD502 pKa = 3.64FDD504 pKa = 4.71GGPLAIAGILLLVGVALLSIGRR526 pKa = 11.84RR527 pKa = 11.84YY528 pKa = 9.93RR529 pKa = 11.84PNGLLRR535 pKa = 11.84PP536 pKa = 4.11
MM1 pKa = 7.56RR2 pKa = 11.84AVKK5 pKa = 9.33RR6 pKa = 11.84TGLAVLASAALGLGALVATPAYY28 pKa = 9.8AATVDD33 pKa = 4.06DD34 pKa = 5.13VIDD37 pKa = 4.81GIIYY41 pKa = 9.89EE42 pKa = 4.68ADD44 pKa = 3.77DD45 pKa = 4.36ADD47 pKa = 4.03VAAGATVTGFDD58 pKa = 3.62GTLTTVNIPDD68 pKa = 3.62TVAIEE73 pKa = 4.02GTEE76 pKa = 3.96YY77 pKa = 11.21DD78 pKa = 3.8VVAIGNSAFVTYY90 pKa = 10.35RR91 pKa = 11.84SGGVITQGDD100 pKa = 3.55ITEE103 pKa = 4.28VVIGDD108 pKa = 3.61NVRR111 pKa = 11.84TIGLQAFMSNRR122 pKa = 11.84IDD124 pKa = 3.74SLTLGSSVTSIKK136 pKa = 10.66GSAFMGNSLTHH147 pKa = 5.25ITIPSAVTSIGGAAFAEE164 pKa = 4.15NDD166 pKa = 3.76DD167 pKa = 4.03LVSVVFTGPAPTVRR181 pKa = 11.84EE182 pKa = 3.7ADD184 pKa = 3.18AGYY187 pKa = 10.76SSASFSTRR195 pKa = 11.84DD196 pKa = 3.27TEE198 pKa = 4.28FVLRR202 pKa = 11.84FPEE205 pKa = 4.5QYY207 pKa = 10.62SADD210 pKa = 3.8NVAGGYY216 pKa = 6.55TTPLWFGYY224 pKa = 5.19TTEE227 pKa = 5.52AIGDD231 pKa = 4.09DD232 pKa = 4.18EE233 pKa = 5.76PDD235 pKa = 3.28ASLSITKK242 pKa = 8.87TVDD245 pKa = 2.68ASFVRR250 pKa = 11.84VYY252 pKa = 11.04DD253 pKa = 3.25WDD255 pKa = 3.9FEE257 pKa = 4.47KK258 pKa = 10.39TGTVGAITVDD268 pKa = 3.41GDD270 pKa = 3.53EE271 pKa = 4.81ASADD275 pKa = 3.24VDD277 pKa = 4.17FTVTATPNGYY287 pKa = 9.43VDD289 pKa = 3.43SYY291 pKa = 11.39YY292 pKa = 11.17VLEE295 pKa = 4.26GTITVLNEE303 pKa = 3.34GTEE306 pKa = 4.11AQTVDD311 pKa = 3.75VVDD314 pKa = 4.37VPDD317 pKa = 3.7VAASASCSVYY327 pKa = 10.74DD328 pKa = 5.1SSFQPAEE335 pKa = 3.99GVEE338 pKa = 4.25IAASEE343 pKa = 4.23SAEE346 pKa = 4.01FEE348 pKa = 4.26YY349 pKa = 10.85SCYY352 pKa = 10.22FDD354 pKa = 3.79GTPEE358 pKa = 4.99DD359 pKa = 4.02GTNTATVTWGDD370 pKa = 3.9DD371 pKa = 3.41QSASATADD379 pKa = 3.31VVFTQRR385 pKa = 11.84DD386 pKa = 3.49TSFQTITIGDD396 pKa = 3.77DD397 pKa = 3.25HH398 pKa = 7.73ADD400 pKa = 3.27PDD402 pKa = 4.6GPFEE406 pKa = 4.12EE407 pKa = 5.75LGTAEE412 pKa = 4.55WNSDD416 pKa = 3.36HH417 pKa = 7.1LPIEE421 pKa = 4.14FTYY424 pKa = 10.86TLTADD429 pKa = 3.49GLTVGEE435 pKa = 4.86CTTLTNTAAIYY446 pKa = 10.05EE447 pKa = 4.34MEE449 pKa = 4.61RR450 pKa = 11.84ATQASVEE457 pKa = 4.24ICPEE461 pKa = 3.92PAVVIPDD468 pKa = 4.33PEE470 pKa = 4.23PTPDD474 pKa = 4.42PEE476 pKa = 4.25PTPDD480 pKa = 4.55PEE482 pKa = 4.47PSPAPVVDD490 pKa = 4.56PEE492 pKa = 4.22PAGLATTGTDD502 pKa = 3.64FDD504 pKa = 4.71GGPLAIAGILLLVGVALLSIGRR526 pKa = 11.84RR527 pKa = 11.84YY528 pKa = 9.93RR529 pKa = 11.84PNGLLRR535 pKa = 11.84PP536 pKa = 4.11
Molecular weight: 55.81 kDa
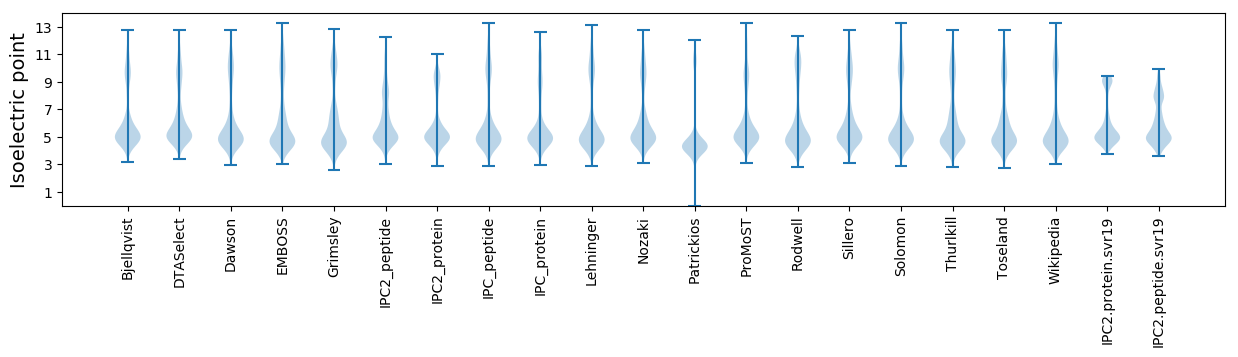
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1R4KAK4|A0A1R4KAK4_9MICO Chromosome (Plasmid) partitioning protein ParA OS=Microbacterium esteraromaticum OX=57043 GN=FM104_11295 PE=4 SV=1
MM1 pKa = 6.49MTTMRR6 pKa = 11.84LFMVMRR12 pKa = 11.84VLARR16 pKa = 11.84TRR18 pKa = 11.84RR19 pKa = 11.84LTSGAMQPLPRR30 pKa = 11.84TRR32 pKa = 11.84ASAVSNSPP40 pKa = 3.11
MM1 pKa = 6.49MTTMRR6 pKa = 11.84LFMVMRR12 pKa = 11.84VLARR16 pKa = 11.84TRR18 pKa = 11.84RR19 pKa = 11.84LTSGAMQPLPRR30 pKa = 11.84TRR32 pKa = 11.84ASAVSNSPP40 pKa = 3.11
Molecular weight: 4.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1007737 |
37 |
2035 |
314.7 |
33.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.275 ± 0.064 | 0.51 ± 0.011 |
6.375 ± 0.037 | 5.771 ± 0.043 |
3.073 ± 0.028 | 8.558 ± 0.035 |
2.099 ± 0.02 | 4.964 ± 0.031 |
2.184 ± 0.033 | 9.932 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.014 ± 0.018 | 2.026 ± 0.024 |
5.169 ± 0.032 | 3.037 ± 0.023 |
7.251 ± 0.054 | 5.767 ± 0.028 |
6.094 ± 0.033 | 8.484 ± 0.044 |
1.496 ± 0.019 | 1.922 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
