
Tursiops truncatus (Atlantic bottle-nosed dolphin) (Delphinus truncatus)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Sarcopterygii; Dipnotetrapodomorpha; Tetrapoda; Amniota; Mammalia; Theria; Eutheria; Boreoeutheria; Laurasiatheria;
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
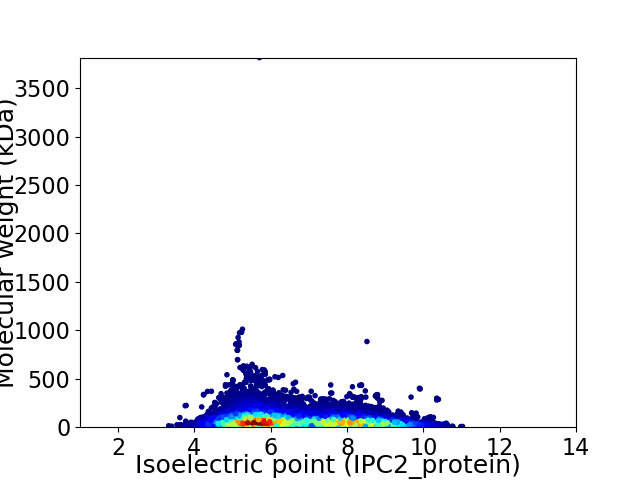
Virtual 2D-PAGE plot for 45130 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6J3PZ46|A0A6J3PZ46_TURTR Isoform of A0A6J3PZC9 transcription elongation factor A protein 2 isoform X2 OS=Tursiops truncatus OX=9739 GN=TCEA2 PE=3 SV=1
MM1 pKa = 7.23EE2 pKa = 5.18AVSPVLFVLGLLLLPLLAVLLMALCVHH29 pKa = 6.24CRR31 pKa = 11.84EE32 pKa = 4.88LPGSYY37 pKa = 8.67DD38 pKa = 3.24TAASDD43 pKa = 3.91GLTPSSIVIKK53 pKa = 9.79PPPTAASWPPATSYY67 pKa = 11.28PPVTYY72 pKa = 10.68LLQSQPDD79 pKa = 3.91LLRR82 pKa = 11.84IPRR85 pKa = 11.84SPQAPGGSPRR95 pKa = 11.84MPSSQQDD102 pKa = 3.01SDD104 pKa = 3.88GANSVASYY112 pKa = 10.11EE113 pKa = 4.13NEE115 pKa = 4.2GASGAPAALAGRR127 pKa = 11.84RR128 pKa = 11.84LGPVLGSADD137 pKa = 3.96PVSLPTPEE145 pKa = 4.78AVCEE149 pKa = 4.13DD150 pKa = 3.73ADD152 pKa = 3.94EE153 pKa = 6.05DD154 pKa = 4.37EE155 pKa = 5.92DD156 pKa = 4.86EE157 pKa = 4.15EE158 pKa = 5.22DD159 pKa = 3.81YY160 pKa = 11.56PEE162 pKa = 5.32GYY164 pKa = 10.61LVVLPDD170 pKa = 3.71NVPATGAAVPPAPASSNPGLRR191 pKa = 11.84DD192 pKa = 3.06SAFSMEE198 pKa = 4.23SGEE201 pKa = 4.99DD202 pKa = 3.63YY203 pKa = 11.66VNVPEE208 pKa = 4.68SEE210 pKa = 4.29EE211 pKa = 4.1SADD214 pKa = 3.58ASLDD218 pKa = 3.44GSRR221 pKa = 11.84EE222 pKa = 3.99YY223 pKa = 11.72VNVSQEE229 pKa = 4.24LPPVARR235 pKa = 11.84TEE237 pKa = 4.35PGVCWGSQPEE247 pKa = 4.17FPGGGGG253 pKa = 3.16
MM1 pKa = 7.23EE2 pKa = 5.18AVSPVLFVLGLLLLPLLAVLLMALCVHH29 pKa = 6.24CRR31 pKa = 11.84EE32 pKa = 4.88LPGSYY37 pKa = 8.67DD38 pKa = 3.24TAASDD43 pKa = 3.91GLTPSSIVIKK53 pKa = 9.79PPPTAASWPPATSYY67 pKa = 11.28PPVTYY72 pKa = 10.68LLQSQPDD79 pKa = 3.91LLRR82 pKa = 11.84IPRR85 pKa = 11.84SPQAPGGSPRR95 pKa = 11.84MPSSQQDD102 pKa = 3.01SDD104 pKa = 3.88GANSVASYY112 pKa = 10.11EE113 pKa = 4.13NEE115 pKa = 4.2GASGAPAALAGRR127 pKa = 11.84RR128 pKa = 11.84LGPVLGSADD137 pKa = 3.96PVSLPTPEE145 pKa = 4.78AVCEE149 pKa = 4.13DD150 pKa = 3.73ADD152 pKa = 3.94EE153 pKa = 6.05DD154 pKa = 4.37EE155 pKa = 5.92DD156 pKa = 4.86EE157 pKa = 4.15EE158 pKa = 5.22DD159 pKa = 3.81YY160 pKa = 11.56PEE162 pKa = 5.32GYY164 pKa = 10.61LVVLPDD170 pKa = 3.71NVPATGAAVPPAPASSNPGLRR191 pKa = 11.84DD192 pKa = 3.06SAFSMEE198 pKa = 4.23SGEE201 pKa = 4.99DD202 pKa = 3.63YY203 pKa = 11.66VNVPEE208 pKa = 4.68SEE210 pKa = 4.29EE211 pKa = 4.1SADD214 pKa = 3.58ASLDD218 pKa = 3.44GSRR221 pKa = 11.84EE222 pKa = 3.99YY223 pKa = 11.72VNVSQEE229 pKa = 4.24LPPVARR235 pKa = 11.84TEE237 pKa = 4.35PGVCWGSQPEE247 pKa = 4.17FPGGGGG253 pKa = 3.16
Molecular weight: 26.09 kDa
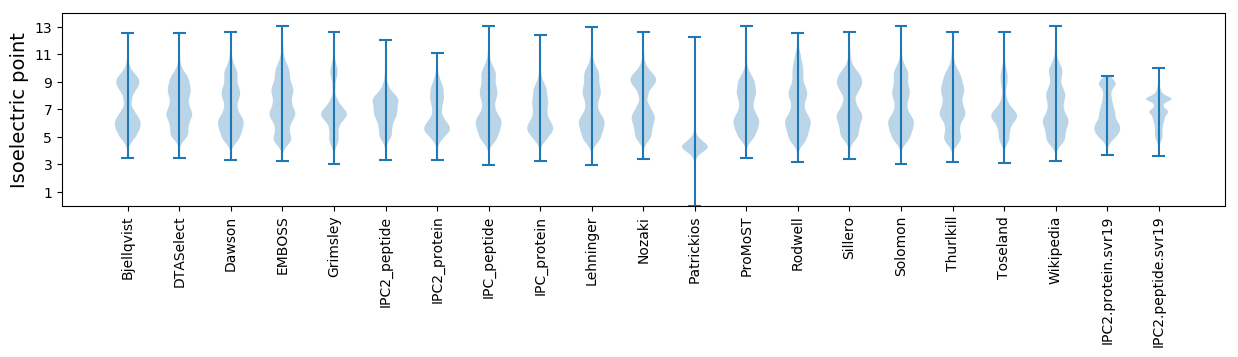
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6J3QL83|A0A6J3QL83_TURTR homeobox protein Hox-B3 OS=Tursiops truncatus OX=9739 GN=HOXB3 PE=3 SV=1
MM1 pKa = 7.4QSISCVLGKK10 pKa = 8.76TPLPRR15 pKa = 11.84QKK17 pKa = 10.47SGSHH21 pKa = 4.93RR22 pKa = 11.84THH24 pKa = 7.41LLTTAHH30 pKa = 6.41SSVKK34 pKa = 10.25SAAKK38 pKa = 9.94SGSHH42 pKa = 5.28RR43 pKa = 11.84THH45 pKa = 7.13LLATAHH51 pKa = 6.3SSVKK55 pKa = 10.42SVAKK59 pKa = 10.2SGSHH63 pKa = 5.01RR64 pKa = 11.84THH66 pKa = 7.13LLATAHH72 pKa = 6.3SSVKK76 pKa = 10.42SVAKK80 pKa = 10.2SGSHH84 pKa = 4.95RR85 pKa = 11.84THH87 pKa = 7.41LLTTAHH93 pKa = 6.41SSVKK97 pKa = 10.25SAAKK101 pKa = 9.94SGSHH105 pKa = 5.28RR106 pKa = 11.84THH108 pKa = 7.13LLATAHH114 pKa = 6.3SSVKK118 pKa = 10.42SVAKK122 pKa = 10.2SGSHH126 pKa = 4.95RR127 pKa = 11.84THH129 pKa = 7.41LLTTAHH135 pKa = 6.43SSVKK139 pKa = 10.36SVAKK143 pKa = 10.2SGSHH147 pKa = 5.01RR148 pKa = 11.84THH150 pKa = 7.13LLATAHH156 pKa = 6.33SSVKK160 pKa = 10.3SAAKK164 pKa = 9.94SGSHH168 pKa = 5.28RR169 pKa = 11.84THH171 pKa = 7.13LLATAHH177 pKa = 6.3SSVKK181 pKa = 10.42SVAKK185 pKa = 10.2SGSHH189 pKa = 4.95RR190 pKa = 11.84THH192 pKa = 7.41LLTTAHH198 pKa = 6.43SSVKK202 pKa = 10.36SVAKK206 pKa = 10.2SGSHH210 pKa = 4.95RR211 pKa = 11.84THH213 pKa = 7.41LLTTAHH219 pKa = 6.43SSVKK223 pKa = 10.36SVAKK227 pKa = 10.2SGSHH231 pKa = 5.01RR232 pKa = 11.84THH234 pKa = 7.13LLATAHH240 pKa = 6.3SSVKK244 pKa = 10.42SVAKK248 pKa = 10.2SGSHH252 pKa = 5.01RR253 pKa = 11.84THH255 pKa = 7.13LLATAHH261 pKa = 6.3SSVKK265 pKa = 10.42SVAKK269 pKa = 10.2SGSHH273 pKa = 4.95RR274 pKa = 11.84THH276 pKa = 7.41LLTTAHH282 pKa = 6.43SSVKK286 pKa = 10.36SVAKK290 pKa = 10.2SGSHH294 pKa = 5.01RR295 pKa = 11.84THH297 pKa = 7.13LLATAHH303 pKa = 6.3SSVKK307 pKa = 10.42SVAKK311 pKa = 10.2SGSHH315 pKa = 4.95RR316 pKa = 11.84THH318 pKa = 7.41LLTTAHH324 pKa = 6.43SSVKK328 pKa = 10.36SVAKK332 pKa = 10.2SGSHH336 pKa = 4.95RR337 pKa = 11.84THH339 pKa = 7.41LLTTAHH345 pKa = 6.43SSVKK349 pKa = 10.32SVAKK353 pKa = 10.24RR354 pKa = 11.84QVEE357 pKa = 3.94DD358 pKa = 3.83SIRR361 pKa = 11.84LTFF364 pKa = 4.2
MM1 pKa = 7.4QSISCVLGKK10 pKa = 8.76TPLPRR15 pKa = 11.84QKK17 pKa = 10.47SGSHH21 pKa = 4.93RR22 pKa = 11.84THH24 pKa = 7.41LLTTAHH30 pKa = 6.41SSVKK34 pKa = 10.25SAAKK38 pKa = 9.94SGSHH42 pKa = 5.28RR43 pKa = 11.84THH45 pKa = 7.13LLATAHH51 pKa = 6.3SSVKK55 pKa = 10.42SVAKK59 pKa = 10.2SGSHH63 pKa = 5.01RR64 pKa = 11.84THH66 pKa = 7.13LLATAHH72 pKa = 6.3SSVKK76 pKa = 10.42SVAKK80 pKa = 10.2SGSHH84 pKa = 4.95RR85 pKa = 11.84THH87 pKa = 7.41LLTTAHH93 pKa = 6.41SSVKK97 pKa = 10.25SAAKK101 pKa = 9.94SGSHH105 pKa = 5.28RR106 pKa = 11.84THH108 pKa = 7.13LLATAHH114 pKa = 6.3SSVKK118 pKa = 10.42SVAKK122 pKa = 10.2SGSHH126 pKa = 4.95RR127 pKa = 11.84THH129 pKa = 7.41LLTTAHH135 pKa = 6.43SSVKK139 pKa = 10.36SVAKK143 pKa = 10.2SGSHH147 pKa = 5.01RR148 pKa = 11.84THH150 pKa = 7.13LLATAHH156 pKa = 6.33SSVKK160 pKa = 10.3SAAKK164 pKa = 9.94SGSHH168 pKa = 5.28RR169 pKa = 11.84THH171 pKa = 7.13LLATAHH177 pKa = 6.3SSVKK181 pKa = 10.42SVAKK185 pKa = 10.2SGSHH189 pKa = 4.95RR190 pKa = 11.84THH192 pKa = 7.41LLTTAHH198 pKa = 6.43SSVKK202 pKa = 10.36SVAKK206 pKa = 10.2SGSHH210 pKa = 4.95RR211 pKa = 11.84THH213 pKa = 7.41LLTTAHH219 pKa = 6.43SSVKK223 pKa = 10.36SVAKK227 pKa = 10.2SGSHH231 pKa = 5.01RR232 pKa = 11.84THH234 pKa = 7.13LLATAHH240 pKa = 6.3SSVKK244 pKa = 10.42SVAKK248 pKa = 10.2SGSHH252 pKa = 5.01RR253 pKa = 11.84THH255 pKa = 7.13LLATAHH261 pKa = 6.3SSVKK265 pKa = 10.42SVAKK269 pKa = 10.2SGSHH273 pKa = 4.95RR274 pKa = 11.84THH276 pKa = 7.41LLTTAHH282 pKa = 6.43SSVKK286 pKa = 10.36SVAKK290 pKa = 10.2SGSHH294 pKa = 5.01RR295 pKa = 11.84THH297 pKa = 7.13LLATAHH303 pKa = 6.3SSVKK307 pKa = 10.42SVAKK311 pKa = 10.2SGSHH315 pKa = 4.95RR316 pKa = 11.84THH318 pKa = 7.41LLTTAHH324 pKa = 6.43SSVKK328 pKa = 10.36SVAKK332 pKa = 10.2SGSHH336 pKa = 4.95RR337 pKa = 11.84THH339 pKa = 7.41LLTTAHH345 pKa = 6.43SSVKK349 pKa = 10.32SVAKK353 pKa = 10.24RR354 pKa = 11.84QVEE357 pKa = 3.94DD358 pKa = 3.83SIRR361 pKa = 11.84LTFF364 pKa = 4.2
Molecular weight: 37.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
31182114 |
28 |
34359 |
690.9 |
76.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.187 ± 0.012 | 2.11 ± 0.009 |
4.845 ± 0.007 | 7.335 ± 0.016 |
3.396 ± 0.008 | 6.584 ± 0.016 |
2.583 ± 0.005 | 4.047 ± 0.01 |
5.712 ± 0.014 | 9.902 ± 0.016 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.047 ± 0.005 | 3.448 ± 0.008 |
6.606 ± 0.021 | 4.99 ± 0.01 |
5.871 ± 0.012 | 8.574 ± 0.012 |
5.213 ± 0.008 | 5.926 ± 0.009 |
1.158 ± 0.004 | 2.454 ± 0.006 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
