
Capybara microvirus Cap3_SP_668
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
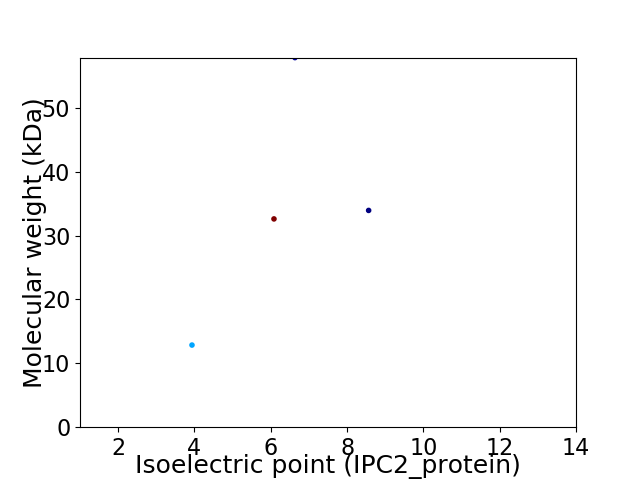
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A4P8W913|A0A4P8W913_9VIRU Uncharacterized protein OS=Capybara microvirus Cap3_SP_668 OX=2585489 PE=4 SV=1
MM1 pKa = 7.29EE2 pKa = 5.49VINIFQYY9 pKa = 10.78VPSEE13 pKa = 4.12GTSFTEE19 pKa = 4.48PSQCIPDD26 pKa = 3.53QTLSVQEE33 pKa = 3.79LLARR37 pKa = 11.84FSRR40 pKa = 11.84GYY42 pKa = 10.51SVDD45 pKa = 3.75VPIHH49 pKa = 7.26DD50 pKa = 5.18DD51 pKa = 3.68YY52 pKa = 11.96EE53 pKa = 4.97SYY55 pKa = 11.03EE56 pKa = 4.15DD57 pKa = 4.46SFEE60 pKa = 3.87EE61 pKa = 4.65DD62 pKa = 4.44AIDD65 pKa = 5.74GIQDD69 pKa = 3.6LSDD72 pKa = 3.23VDD74 pKa = 3.42ILRR77 pKa = 11.84EE78 pKa = 3.91QIHH81 pKa = 5.24MQKK84 pKa = 10.71KK85 pKa = 9.0LAQTSDD91 pKa = 3.27VMISEE96 pKa = 4.42VTDD99 pKa = 3.6KK100 pKa = 10.92EE101 pKa = 4.15RR102 pKa = 11.84SDD104 pKa = 3.97AKK106 pKa = 11.33GNDD109 pKa = 3.45EE110 pKa = 4.67SSASS114 pKa = 3.49
MM1 pKa = 7.29EE2 pKa = 5.49VINIFQYY9 pKa = 10.78VPSEE13 pKa = 4.12GTSFTEE19 pKa = 4.48PSQCIPDD26 pKa = 3.53QTLSVQEE33 pKa = 3.79LLARR37 pKa = 11.84FSRR40 pKa = 11.84GYY42 pKa = 10.51SVDD45 pKa = 3.75VPIHH49 pKa = 7.26DD50 pKa = 5.18DD51 pKa = 3.68YY52 pKa = 11.96EE53 pKa = 4.97SYY55 pKa = 11.03EE56 pKa = 4.15DD57 pKa = 4.46SFEE60 pKa = 3.87EE61 pKa = 4.65DD62 pKa = 4.44AIDD65 pKa = 5.74GIQDD69 pKa = 3.6LSDD72 pKa = 3.23VDD74 pKa = 3.42ILRR77 pKa = 11.84EE78 pKa = 3.91QIHH81 pKa = 5.24MQKK84 pKa = 10.71KK85 pKa = 9.0LAQTSDD91 pKa = 3.27VMISEE96 pKa = 4.42VTDD99 pKa = 3.6KK100 pKa = 10.92EE101 pKa = 4.15RR102 pKa = 11.84SDD104 pKa = 3.97AKK106 pKa = 11.33GNDD109 pKa = 3.45EE110 pKa = 4.67SSASS114 pKa = 3.49
Molecular weight: 12.86 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W5F6|A0A4P8W5F6_9VIRU Major capsid protein OS=Capybara microvirus Cap3_SP_668 OX=2585489 PE=3 SV=1
MM1 pKa = 7.39KK2 pKa = 10.02CVSPVTIRR10 pKa = 11.84NPNFGNISKK19 pKa = 10.13IGYY22 pKa = 8.06DD23 pKa = 3.02SMYY26 pKa = 10.49IGVPCGKK33 pKa = 9.91CLPCLQNKK41 pKa = 9.26RR42 pKa = 11.84NDD44 pKa = 2.8WYY46 pKa = 11.36LRR48 pKa = 11.84LLSEE52 pKa = 4.54YY53 pKa = 10.32RR54 pKa = 11.84SSTNSCFVTLTYY66 pKa = 10.75SDD68 pKa = 4.56EE69 pKa = 4.41NCDD72 pKa = 3.48GSVHH76 pKa = 6.63KK77 pKa = 10.69KK78 pKa = 10.49DD79 pKa = 3.14IQEE82 pKa = 3.8FFARR86 pKa = 11.84LRR88 pKa = 11.84KK89 pKa = 9.44RR90 pKa = 11.84LQYY93 pKa = 10.93HH94 pKa = 7.0LGTVGLRR101 pKa = 11.84FFCVSEE107 pKa = 4.15YY108 pKa = 11.02GPKK111 pKa = 7.62THH113 pKa = 7.15RR114 pKa = 11.84PHH116 pKa = 5.56YY117 pKa = 7.76HH118 pKa = 5.32TLIFGYY124 pKa = 9.17PDD126 pKa = 3.93CDD128 pKa = 3.21INHH131 pKa = 6.67YY132 pKa = 9.92IEE134 pKa = 4.58KK135 pKa = 9.66SWYY138 pKa = 9.88LGFVCVTNININRR151 pKa = 11.84IRR153 pKa = 11.84YY154 pKa = 6.21CAKK157 pKa = 10.09YY158 pKa = 10.05CISKK162 pKa = 10.73NDD164 pKa = 3.34VPEE167 pKa = 4.56NMDD170 pKa = 4.04NNFMLCSRR178 pKa = 11.84RR179 pKa = 11.84PAIGDD184 pKa = 3.49SFLTNKK190 pKa = 9.39VKK192 pKa = 10.5SFYY195 pKa = 10.62SKK197 pKa = 9.98RR198 pKa = 11.84HH199 pKa = 4.62GKK201 pKa = 9.75IIYY204 pKa = 9.1NDD206 pKa = 3.28HH207 pKa = 5.83GVNYY211 pKa = 10.1SLPRR215 pKa = 11.84RR216 pKa = 11.84WKK218 pKa = 10.4EE219 pKa = 3.53KK220 pKa = 10.68LLSEE224 pKa = 4.14KK225 pKa = 10.39QKK227 pKa = 11.41NEE229 pKa = 3.49IKK231 pKa = 10.13HH232 pKa = 5.97NSMIYY237 pKa = 10.96NNDD240 pKa = 3.43CYY242 pKa = 11.13KK243 pKa = 10.74KK244 pKa = 9.7YY245 pKa = 9.21QEE247 pKa = 4.88KK248 pKa = 9.9YY249 pKa = 9.31AAYY252 pKa = 10.22DD253 pKa = 3.62EE254 pKa = 4.92AALAHH259 pKa = 6.48GNPIMSQLQIDD270 pKa = 3.96QKK272 pKa = 10.98LEE274 pKa = 4.05SMKK277 pKa = 10.67KK278 pKa = 10.44YY279 pKa = 10.38LDD281 pKa = 3.69KK282 pKa = 10.58DD283 pKa = 3.87TKK285 pKa = 10.86FKK287 pKa = 11.17LL288 pKa = 3.56
MM1 pKa = 7.39KK2 pKa = 10.02CVSPVTIRR10 pKa = 11.84NPNFGNISKK19 pKa = 10.13IGYY22 pKa = 8.06DD23 pKa = 3.02SMYY26 pKa = 10.49IGVPCGKK33 pKa = 9.91CLPCLQNKK41 pKa = 9.26RR42 pKa = 11.84NDD44 pKa = 2.8WYY46 pKa = 11.36LRR48 pKa = 11.84LLSEE52 pKa = 4.54YY53 pKa = 10.32RR54 pKa = 11.84SSTNSCFVTLTYY66 pKa = 10.75SDD68 pKa = 4.56EE69 pKa = 4.41NCDD72 pKa = 3.48GSVHH76 pKa = 6.63KK77 pKa = 10.69KK78 pKa = 10.49DD79 pKa = 3.14IQEE82 pKa = 3.8FFARR86 pKa = 11.84LRR88 pKa = 11.84KK89 pKa = 9.44RR90 pKa = 11.84LQYY93 pKa = 10.93HH94 pKa = 7.0LGTVGLRR101 pKa = 11.84FFCVSEE107 pKa = 4.15YY108 pKa = 11.02GPKK111 pKa = 7.62THH113 pKa = 7.15RR114 pKa = 11.84PHH116 pKa = 5.56YY117 pKa = 7.76HH118 pKa = 5.32TLIFGYY124 pKa = 9.17PDD126 pKa = 3.93CDD128 pKa = 3.21INHH131 pKa = 6.67YY132 pKa = 9.92IEE134 pKa = 4.58KK135 pKa = 9.66SWYY138 pKa = 9.88LGFVCVTNININRR151 pKa = 11.84IRR153 pKa = 11.84YY154 pKa = 6.21CAKK157 pKa = 10.09YY158 pKa = 10.05CISKK162 pKa = 10.73NDD164 pKa = 3.34VPEE167 pKa = 4.56NMDD170 pKa = 4.04NNFMLCSRR178 pKa = 11.84RR179 pKa = 11.84PAIGDD184 pKa = 3.49SFLTNKK190 pKa = 9.39VKK192 pKa = 10.5SFYY195 pKa = 10.62SKK197 pKa = 9.98RR198 pKa = 11.84HH199 pKa = 4.62GKK201 pKa = 9.75IIYY204 pKa = 9.1NDD206 pKa = 3.28HH207 pKa = 5.83GVNYY211 pKa = 10.1SLPRR215 pKa = 11.84RR216 pKa = 11.84WKK218 pKa = 10.4EE219 pKa = 3.53KK220 pKa = 10.68LLSEE224 pKa = 4.14KK225 pKa = 10.39QKK227 pKa = 11.41NEE229 pKa = 3.49IKK231 pKa = 10.13HH232 pKa = 5.97NSMIYY237 pKa = 10.96NNDD240 pKa = 3.43CYY242 pKa = 11.13KK243 pKa = 10.74KK244 pKa = 9.7YY245 pKa = 9.21QEE247 pKa = 4.88KK248 pKa = 9.9YY249 pKa = 9.31AAYY252 pKa = 10.22DD253 pKa = 3.62EE254 pKa = 4.92AALAHH259 pKa = 6.48GNPIMSQLQIDD270 pKa = 3.96QKK272 pKa = 10.98LEE274 pKa = 4.05SMKK277 pKa = 10.67KK278 pKa = 10.44YY279 pKa = 10.38LDD281 pKa = 3.69KK282 pKa = 10.58DD283 pKa = 3.87TKK285 pKa = 10.86FKK287 pKa = 11.17LL288 pKa = 3.56
Molecular weight: 33.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1203 |
114 |
516 |
300.8 |
34.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.403 ± 0.906 | 1.413 ± 0.872 |
5.653 ± 1.094 | 5.902 ± 0.941 |
3.99 ± 0.66 | 5.819 ± 1.029 |
2.078 ± 0.42 | 5.902 ± 0.693 |
6.733 ± 1.097 | 8.562 ± 1.085 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.159 ± 0.724 | 5.569 ± 1.077 |
4.572 ± 0.647 | 5.653 ± 1.0 |
4.655 ± 0.443 | 9.476 ± 0.934 |
4.406 ± 0.431 | 4.572 ± 0.459 |
1.164 ± 0.254 | 5.32 ± 0.693 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
