
Candidatus Methylopumilus turicensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Nitrosomonadales; Methylophilaceae; Candidatus Methylopumilus
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
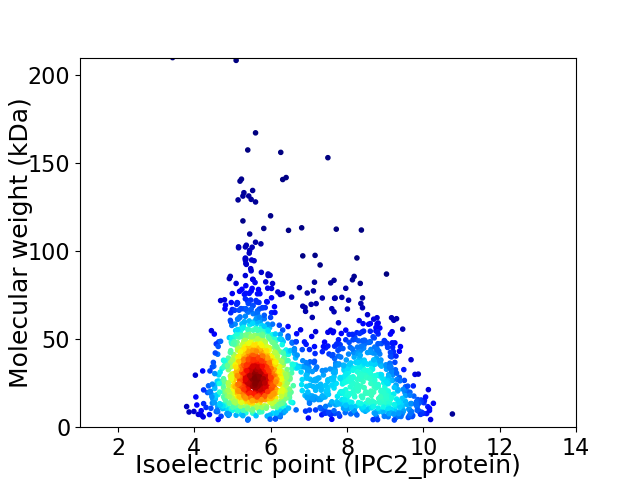
Virtual 2D-PAGE plot for 1739 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
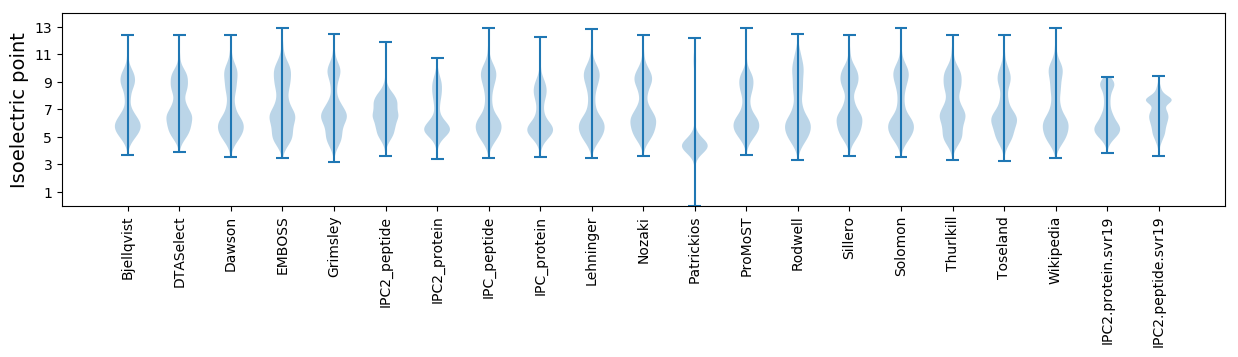
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B7IVB6|A0A0B7IVB6_9PROT Uncharacterized protein OS=Candidatus Methylopumilus turicensis OX=1581680 GN=BN1209_1109 PE=4 SV=1
MM1 pKa = 7.26ATKK4 pKa = 10.44KK5 pKa = 10.4PQDD8 pKa = 4.02DD9 pKa = 3.95VTKK12 pKa = 10.76VPVQQGVDD20 pKa = 3.29AEE22 pKa = 4.49STYY25 pKa = 11.22VAAQTANPDD34 pKa = 3.97KK35 pKa = 11.09QWMDD39 pKa = 3.64DD40 pKa = 3.42TAKK43 pKa = 10.74AWDD46 pKa = 3.61ARR48 pKa = 11.84KK49 pKa = 9.65SAEE52 pKa = 4.23KK53 pKa = 10.58EE54 pKa = 3.59NDD56 pKa = 3.37VVKK59 pKa = 11.23VEE61 pKa = 4.12GQAVINLADD70 pKa = 3.35ATGAVGEE77 pKa = 4.39KK78 pKa = 9.47TGAALIVADD87 pKa = 4.16TANSKK92 pKa = 10.42DD93 pKa = 3.31AAVVVKK99 pKa = 10.8KK100 pKa = 10.73KK101 pKa = 10.73DD102 pKa = 3.62DD103 pKa = 4.21DD104 pKa = 3.93SHH106 pKa = 8.06DD107 pKa = 3.34KK108 pKa = 11.16GGYY111 pKa = 7.4IWGGLGVLGLIGIAAAGGGGGGDD134 pKa = 3.79SAPAPAPAPLRR145 pKa = 11.84LDD147 pKa = 3.51GLVIDD152 pKa = 5.45DD153 pKa = 4.57PVANAIVFRR162 pKa = 11.84DD163 pKa = 4.09TYY165 pKa = 11.11GDD167 pKa = 3.99GYY169 pKa = 10.86LHH171 pKa = 6.94VIDD174 pKa = 5.17AGEE177 pKa = 3.73GRR179 pKa = 11.84YY180 pKa = 9.26IVDD183 pKa = 3.68PDD185 pKa = 3.36QSVVVTNALGEE196 pKa = 4.05FSSLGGSGRR205 pKa = 11.84IISVGINRR213 pKa = 11.84LPILVHH219 pKa = 6.85EE220 pKa = 4.99DD221 pKa = 3.77AEE223 pKa = 4.41DD224 pKa = 3.55LTPQEE229 pKa = 4.2MVANEE234 pKa = 4.04GDD236 pKa = 4.08LLFASAAKK244 pKa = 9.8EE245 pKa = 4.04VANVAINNGDD255 pKa = 3.44YY256 pKa = 11.39AFIVGEE262 pKa = 3.96NQIFDD267 pKa = 4.05SLVISSGITLSVGGANVTLVTGVTALVDD295 pKa = 4.26AYY297 pKa = 9.62IHH299 pKa = 6.94RR300 pKa = 11.84YY301 pKa = 8.99EE302 pKa = 5.28DD303 pKa = 4.13DD304 pKa = 3.67EE305 pKa = 4.48FVPTNAEE312 pKa = 3.69VKK314 pKa = 10.25AAIIEE319 pKa = 4.2VANLFGYY326 pKa = 9.02PLNAGSSQQALMSFLLGGYY345 pKa = 9.54DD346 pKa = 3.45VAGVNSPDD354 pKa = 3.61NLNQEE359 pKa = 4.21IYY361 pKa = 10.79SIVNSLYY368 pKa = 10.04ATNEE372 pKa = 3.76ALNDD376 pKa = 3.95DD377 pKa = 4.42QYY379 pKa = 12.2DD380 pKa = 3.61AADD383 pKa = 3.41YY384 pKa = 11.93SMVFNNALNAIILVQEE400 pKa = 3.75EE401 pKa = 4.2RR402 pKa = 11.84VRR404 pKa = 11.84NDD406 pKa = 3.38EE407 pKa = 4.14EE408 pKa = 5.04DD409 pKa = 3.84LASLATPEE417 pKa = 3.91VFANVTEE424 pKa = 4.39EE425 pKa = 4.65FIQLFNQISDD435 pKa = 3.51NDD437 pKa = 3.31ISTNEE442 pKa = 3.72AQLFIDD448 pKa = 4.01VASNHH453 pKa = 6.25ANFGGGAADD462 pKa = 2.95IYY464 pKa = 10.26EE465 pKa = 4.61TFDD468 pKa = 3.13EE469 pKa = 5.25SYY471 pKa = 9.66STDD474 pKa = 3.55DD475 pKa = 3.94DD476 pKa = 4.25NLVNMVVGGDD486 pKa = 3.42QLRR489 pKa = 11.84EE490 pKa = 3.67QDD492 pKa = 3.76VEE494 pKa = 4.51VVGNITVLAAGTNDD508 pKa = 3.57SVDD511 pKa = 4.47LILFGGEE518 pKa = 3.99DD519 pKa = 3.81SPVEE523 pKa = 3.82LMGSNILVSAYY534 pKa = 7.55GTSSSADD541 pKa = 3.27LVLAAVNAPGTEE553 pKa = 4.12MSNGTYY559 pKa = 9.86TYY561 pKa = 10.58DD562 pKa = 3.25ARR564 pKa = 11.84SDD566 pKa = 3.4EE567 pKa = 4.42TVAVGDD573 pKa = 3.65ITVQATGEE581 pKa = 4.02DD582 pKa = 4.41SEE584 pKa = 5.08SGLYY588 pKa = 9.49MSADD592 pKa = 3.64LDD594 pKa = 3.64IAGDD598 pKa = 3.68VSITSSGEE606 pKa = 3.66DD607 pKa = 3.2SSAVAQMISIAGMAQSAIFEE627 pKa = 4.2VDD629 pKa = 3.42LAAIIEE635 pKa = 4.49DD636 pKa = 3.83ALTTDD641 pKa = 3.43RR642 pKa = 11.84SVIQLVVDD650 pKa = 4.45GKK652 pKa = 9.09VLRR655 pKa = 11.84ANVTEE660 pKa = 4.21GDD662 pKa = 3.51TDD664 pKa = 3.59EE665 pKa = 4.4VVANIIEE672 pKa = 4.16QWKK675 pKa = 10.19LDD677 pKa = 3.53SDD679 pKa = 4.32YY680 pKa = 11.29STLSEE685 pKa = 4.0NQMEE689 pKa = 4.46YY690 pKa = 9.21WGDD693 pKa = 3.37YY694 pKa = 10.41ARR696 pKa = 11.84IRR698 pKa = 11.84FDD700 pKa = 3.03NTEE703 pKa = 3.76SHH705 pKa = 6.15EE706 pKa = 4.42VSVSRR711 pKa = 11.84YY712 pKa = 8.64NGPIEE717 pKa = 3.91TDD719 pKa = 3.52FDD721 pKa = 4.34VVGTVQDD728 pKa = 3.42NYY730 pKa = 10.7EE731 pKa = 3.98RR732 pKa = 11.84EE733 pKa = 3.78AAINFEE739 pKa = 4.13GSHH742 pKa = 7.49IEE744 pKa = 4.13VQALGNNFFNNNGSYY759 pKa = 9.32DD760 pKa = 3.44TDD762 pKa = 3.44AALVVFGATGSINGIDD778 pKa = 4.03VIAGGNDD785 pKa = 3.13YY786 pKa = 10.84FGNYY790 pKa = 10.01SFDD793 pKa = 3.89LPNALAIINIDD804 pKa = 3.38GSVSGEE810 pKa = 3.9INVTTLEE817 pKa = 4.17EE818 pKa = 4.41GASKK822 pKa = 10.0SAYY825 pKa = 9.32MLLQATAGEE834 pKa = 4.33LSLDD838 pKa = 3.52DD839 pKa = 3.39VTINVTAAGNNSSADD854 pKa = 3.75LEE856 pKa = 4.56IGGDD860 pKa = 4.07LNNLPIPGFWSTDD873 pKa = 3.25SVNTLVTGSINGITVSASGDD893 pKa = 3.5NSDD896 pKa = 4.4ASLDD900 pKa = 3.43ISGDD904 pKa = 2.92ITIRR908 pKa = 11.84GTILVEE914 pKa = 4.44TSAEE918 pKa = 3.95DD919 pKa = 3.49ANAEE923 pKa = 4.11LSIDD927 pKa = 4.05SNFDD931 pKa = 3.29SSDD934 pKa = 3.54TLSSSVITFDD944 pKa = 3.52NVDD947 pKa = 3.91IIARR951 pKa = 11.84AYY953 pKa = 10.46GEE955 pKa = 4.26SSDD958 pKa = 3.83VDD960 pKa = 3.23ISIDD964 pKa = 3.27GNAYY968 pKa = 10.32GSINSILLVAHH979 pKa = 7.38ADD981 pKa = 3.61GALDD985 pKa = 3.81GSRR988 pKa = 11.84NAEE991 pKa = 4.05SADD994 pKa = 3.62LNVDD998 pKa = 3.08INFNGTVGDD1007 pKa = 3.59ITLQAMGNSDD1017 pKa = 5.66DD1018 pKa = 4.71VDD1020 pKa = 5.32LDD1022 pKa = 3.35MDD1024 pKa = 4.37ASSNRR1029 pKa = 11.84SNGDD1033 pKa = 2.92EE1034 pKa = 4.07VQFYY1038 pKa = 11.19DD1039 pKa = 4.15LGQMMSYY1046 pKa = 10.72DD1047 pKa = 4.32SSWQDD1052 pKa = 3.37LQTGDD1057 pKa = 5.17LISLTIGDD1065 pKa = 4.26VVLTATVGQDD1075 pKa = 3.07VNDD1078 pKa = 4.55DD1079 pKa = 3.52YY1080 pKa = 11.52GTAVEE1085 pKa = 4.31VLADD1089 pKa = 3.81VVSQLQDD1096 pKa = 2.99AVDD1099 pKa = 3.82ARR1101 pKa = 11.84NVDD1104 pKa = 3.7PDD1106 pKa = 4.04LSDD1109 pKa = 3.99LNVVIKK1115 pKa = 8.66QTGNTNYY1122 pKa = 9.92FDD1124 pKa = 3.9NYY1126 pKa = 10.42YY1127 pKa = 10.26GADD1130 pKa = 4.39GIWVRR1135 pKa = 11.84WLDD1138 pKa = 3.67AGEE1141 pKa = 4.17QEE1143 pKa = 4.28ATSLNVDD1150 pKa = 3.5GDD1152 pKa = 4.02IVSPTDD1158 pKa = 3.25RR1159 pKa = 11.84EE1160 pKa = 4.35YY1161 pKa = 11.68GNDD1164 pKa = 3.34MPTSIEE1170 pKa = 3.99FLDD1173 pKa = 3.93SNILIEE1179 pKa = 4.17AHH1181 pKa = 6.46GEE1183 pKa = 4.08SSDD1186 pKa = 4.41ASLDD1190 pKa = 3.74VNDD1193 pKa = 3.4ATGRR1197 pKa = 11.84INSLTLIATDD1207 pKa = 3.98EE1208 pKa = 4.46NGGSADD1214 pKa = 3.13VDD1216 pKa = 3.55ADD1218 pKa = 3.55IEE1220 pKa = 4.4LGGRR1224 pKa = 11.84LGDD1227 pKa = 3.61VTITADD1233 pKa = 3.76GYY1235 pKa = 8.51NTEE1238 pKa = 4.68AQVEE1242 pKa = 4.52MNSAPEE1248 pKa = 3.9GLTYY1252 pKa = 10.91DD1253 pKa = 3.95NSIITLNALGDD1264 pKa = 4.05SADD1267 pKa = 3.66VEE1269 pKa = 4.61YY1270 pKa = 10.69HH1271 pKa = 5.78QEE1273 pKa = 4.08DD1274 pKa = 4.47YY1275 pKa = 11.53GQGTIDD1281 pKa = 3.55SLNINVIGNNYY1292 pKa = 9.78ADD1294 pKa = 4.14ADD1296 pKa = 4.23VIIGGDD1302 pKa = 3.02ITFDD1306 pKa = 4.11SEE1308 pKa = 3.93INLYY1312 pKa = 10.27ARR1314 pKa = 11.84NTEE1317 pKa = 3.87SSDD1320 pKa = 3.21GVNIYY1325 pKa = 8.64MHH1327 pKa = 7.32LANEE1331 pKa = 4.32DD1332 pKa = 3.46RR1333 pKa = 11.84DD1334 pKa = 4.18DD1335 pKa = 4.0GMNSTFHH1342 pKa = 6.44MNDD1345 pKa = 2.41VTINVEE1351 pKa = 4.24SVGYY1355 pKa = 8.95NARR1358 pKa = 11.84AEE1360 pKa = 4.18ITTQNYY1366 pKa = 9.22AHH1368 pKa = 7.12TDD1370 pKa = 3.18GDD1372 pKa = 4.02FSSDD1376 pKa = 3.19SSIDD1380 pKa = 3.39STITGTINALNVSAGVYY1397 pKa = 10.16DD1398 pKa = 5.25DD1399 pKa = 4.58SNEE1402 pKa = 4.49SINPTMVANAEE1413 pKa = 4.2SSVDD1417 pKa = 3.95LNVHH1421 pKa = 6.63GIIEE1425 pKa = 4.99SITLVADD1432 pKa = 3.5NVSEE1436 pKa = 4.23VSVDD1440 pKa = 3.45LFGAINANGVMDD1452 pKa = 3.65ITITNSNSSSISLDD1466 pKa = 3.33LYY1468 pKa = 11.08DD1469 pKa = 3.75VYY1471 pKa = 11.69GDD1473 pKa = 3.83IEE1475 pKa = 5.0TITSNSGSEE1484 pKa = 4.01GAEE1487 pKa = 3.76TNIYY1491 pKa = 10.46VSTEE1495 pKa = 3.68GSIQTVNLTTTQDD1508 pKa = 3.4AEE1510 pKa = 4.65TVASFDD1516 pKa = 3.82INGSQHH1522 pKa = 6.56YY1523 pKa = 8.96INNMTITTVGTSFFGDD1539 pKa = 3.63YY1540 pKa = 10.52TSVSDD1545 pKa = 4.75VIVDD1549 pKa = 3.69QEE1551 pKa = 4.08AHH1553 pKa = 6.09GGVVNIVNGTSNGDD1567 pKa = 3.59LLADD1571 pKa = 3.87TRR1573 pKa = 11.84LSYY1576 pKa = 10.57YY1577 pKa = 11.02GNYY1580 pKa = 9.05QADD1583 pKa = 4.3SIVIAANNNDD1593 pKa = 3.42GAGDD1597 pKa = 3.72EE1598 pKa = 4.43FALHH1602 pKa = 6.81ISMDD1606 pKa = 3.93DD1607 pKa = 3.17VDD1609 pKa = 4.88VYY1611 pKa = 10.63STQHH1615 pKa = 4.36QTALVNNMTSITGLVNGYY1633 pKa = 10.41DD1634 pKa = 3.37NPTNNFIQFADD1645 pKa = 4.65LEE1647 pKa = 5.07DD1648 pKa = 4.95DD1649 pKa = 4.42DD1650 pKa = 4.4YY1651 pKa = 11.66TYY1653 pKa = 11.38EE1654 pKa = 4.38PGANGRR1660 pKa = 11.84QGPIDD1665 pKa = 4.55LNISHH1670 pKa = 6.84YY1671 pKa = 11.49SNGLDD1676 pKa = 3.07GWVGGDD1682 pKa = 3.67TYY1684 pKa = 11.4FDD1686 pKa = 4.83DD1687 pKa = 5.32YY1688 pKa = 11.52EE1689 pKa = 5.04GSQNIGNGFYY1699 pKa = 11.04GSFNEE1704 pKa = 5.12FFDD1707 pKa = 3.82AAQFGIASEE1716 pKa = 4.39YY1717 pKa = 11.32AFVEE1721 pKa = 4.46LSQYY1725 pKa = 9.89GAAGHH1730 pKa = 7.01DD1731 pKa = 4.05GYY1733 pKa = 11.44DD1734 pKa = 3.24FGFDD1738 pKa = 2.84TDD1740 pKa = 3.66VYY1742 pKa = 11.41LNVSWVDD1749 pKa = 3.43KK1750 pKa = 10.91NGNSQTIYY1758 pKa = 11.05LNADD1762 pKa = 3.47LGGSSLGGSLAGDD1775 pKa = 3.65ATAVVNGLNADD1786 pKa = 4.01TDD1788 pKa = 3.72WDD1790 pKa = 4.01GTPTGSLDD1798 pKa = 3.71GSDD1801 pKa = 4.26GRR1803 pKa = 11.84WDD1805 pKa = 3.25SWDD1808 pKa = 3.42NNLTVFANDD1817 pKa = 3.34SNTLGIRR1824 pKa = 11.84IGEE1827 pKa = 4.27GYY1829 pKa = 7.54ITNVSFHH1836 pKa = 6.9SDD1838 pKa = 2.58WVEE1841 pKa = 3.79VDD1843 pKa = 5.01DD1844 pKa = 6.05LVNDD1848 pKa = 3.64YY1849 pKa = 10.9TNIGSSDD1856 pKa = 3.13ASQSKK1861 pKa = 10.62GYY1863 pKa = 9.29TSLNDD1868 pKa = 3.2YY1869 pKa = 11.7ANFEE1873 pKa = 4.13EE1874 pKa = 5.24SSIQFVEE1881 pKa = 4.09GGGYY1885 pKa = 9.81FFGMVLNEE1893 pKa = 4.29DD1894 pKa = 3.57QNSYY1898 pKa = 10.83RR1899 pKa = 11.84GVLAMDD1905 pKa = 3.89EE1906 pKa = 4.54DD1907 pKa = 5.01GEE1909 pKa = 4.82GVTKK1913 pKa = 10.76LIEE1916 pKa = 4.29LVDD1919 pKa = 4.04VYY1921 pKa = 11.56ASYY1924 pKa = 11.9NDD1926 pKa = 2.72VDD1928 pKa = 3.94YY1929 pKa = 11.49YY1930 pKa = 11.55LNEE1933 pKa = 3.96GDD1935 pKa = 5.11ADD1937 pKa = 3.44HH1938 pKa = 7.27GYY1940 pKa = 11.21NDD1942 pKa = 4.15FSSAYY1947 pKa = 9.68IGMGYY1952 pKa = 10.31GNEE1955 pKa = 4.19GGGFAGYY1962 pKa = 10.34AGSTGFDD1969 pKa = 3.66YY1970 pKa = 8.1EE1971 pKa = 4.55TNADD1975 pKa = 3.18NYY1977 pKa = 11.19AFVVV1981 pKa = 3.57
MM1 pKa = 7.26ATKK4 pKa = 10.44KK5 pKa = 10.4PQDD8 pKa = 4.02DD9 pKa = 3.95VTKK12 pKa = 10.76VPVQQGVDD20 pKa = 3.29AEE22 pKa = 4.49STYY25 pKa = 11.22VAAQTANPDD34 pKa = 3.97KK35 pKa = 11.09QWMDD39 pKa = 3.64DD40 pKa = 3.42TAKK43 pKa = 10.74AWDD46 pKa = 3.61ARR48 pKa = 11.84KK49 pKa = 9.65SAEE52 pKa = 4.23KK53 pKa = 10.58EE54 pKa = 3.59NDD56 pKa = 3.37VVKK59 pKa = 11.23VEE61 pKa = 4.12GQAVINLADD70 pKa = 3.35ATGAVGEE77 pKa = 4.39KK78 pKa = 9.47TGAALIVADD87 pKa = 4.16TANSKK92 pKa = 10.42DD93 pKa = 3.31AAVVVKK99 pKa = 10.8KK100 pKa = 10.73KK101 pKa = 10.73DD102 pKa = 3.62DD103 pKa = 4.21DD104 pKa = 3.93SHH106 pKa = 8.06DD107 pKa = 3.34KK108 pKa = 11.16GGYY111 pKa = 7.4IWGGLGVLGLIGIAAAGGGGGGDD134 pKa = 3.79SAPAPAPAPLRR145 pKa = 11.84LDD147 pKa = 3.51GLVIDD152 pKa = 5.45DD153 pKa = 4.57PVANAIVFRR162 pKa = 11.84DD163 pKa = 4.09TYY165 pKa = 11.11GDD167 pKa = 3.99GYY169 pKa = 10.86LHH171 pKa = 6.94VIDD174 pKa = 5.17AGEE177 pKa = 3.73GRR179 pKa = 11.84YY180 pKa = 9.26IVDD183 pKa = 3.68PDD185 pKa = 3.36QSVVVTNALGEE196 pKa = 4.05FSSLGGSGRR205 pKa = 11.84IISVGINRR213 pKa = 11.84LPILVHH219 pKa = 6.85EE220 pKa = 4.99DD221 pKa = 3.77AEE223 pKa = 4.41DD224 pKa = 3.55LTPQEE229 pKa = 4.2MVANEE234 pKa = 4.04GDD236 pKa = 4.08LLFASAAKK244 pKa = 9.8EE245 pKa = 4.04VANVAINNGDD255 pKa = 3.44YY256 pKa = 11.39AFIVGEE262 pKa = 3.96NQIFDD267 pKa = 4.05SLVISSGITLSVGGANVTLVTGVTALVDD295 pKa = 4.26AYY297 pKa = 9.62IHH299 pKa = 6.94RR300 pKa = 11.84YY301 pKa = 8.99EE302 pKa = 5.28DD303 pKa = 4.13DD304 pKa = 3.67EE305 pKa = 4.48FVPTNAEE312 pKa = 3.69VKK314 pKa = 10.25AAIIEE319 pKa = 4.2VANLFGYY326 pKa = 9.02PLNAGSSQQALMSFLLGGYY345 pKa = 9.54DD346 pKa = 3.45VAGVNSPDD354 pKa = 3.61NLNQEE359 pKa = 4.21IYY361 pKa = 10.79SIVNSLYY368 pKa = 10.04ATNEE372 pKa = 3.76ALNDD376 pKa = 3.95DD377 pKa = 4.42QYY379 pKa = 12.2DD380 pKa = 3.61AADD383 pKa = 3.41YY384 pKa = 11.93SMVFNNALNAIILVQEE400 pKa = 3.75EE401 pKa = 4.2RR402 pKa = 11.84VRR404 pKa = 11.84NDD406 pKa = 3.38EE407 pKa = 4.14EE408 pKa = 5.04DD409 pKa = 3.84LASLATPEE417 pKa = 3.91VFANVTEE424 pKa = 4.39EE425 pKa = 4.65FIQLFNQISDD435 pKa = 3.51NDD437 pKa = 3.31ISTNEE442 pKa = 3.72AQLFIDD448 pKa = 4.01VASNHH453 pKa = 6.25ANFGGGAADD462 pKa = 2.95IYY464 pKa = 10.26EE465 pKa = 4.61TFDD468 pKa = 3.13EE469 pKa = 5.25SYY471 pKa = 9.66STDD474 pKa = 3.55DD475 pKa = 3.94DD476 pKa = 4.25NLVNMVVGGDD486 pKa = 3.42QLRR489 pKa = 11.84EE490 pKa = 3.67QDD492 pKa = 3.76VEE494 pKa = 4.51VVGNITVLAAGTNDD508 pKa = 3.57SVDD511 pKa = 4.47LILFGGEE518 pKa = 3.99DD519 pKa = 3.81SPVEE523 pKa = 3.82LMGSNILVSAYY534 pKa = 7.55GTSSSADD541 pKa = 3.27LVLAAVNAPGTEE553 pKa = 4.12MSNGTYY559 pKa = 9.86TYY561 pKa = 10.58DD562 pKa = 3.25ARR564 pKa = 11.84SDD566 pKa = 3.4EE567 pKa = 4.42TVAVGDD573 pKa = 3.65ITVQATGEE581 pKa = 4.02DD582 pKa = 4.41SEE584 pKa = 5.08SGLYY588 pKa = 9.49MSADD592 pKa = 3.64LDD594 pKa = 3.64IAGDD598 pKa = 3.68VSITSSGEE606 pKa = 3.66DD607 pKa = 3.2SSAVAQMISIAGMAQSAIFEE627 pKa = 4.2VDD629 pKa = 3.42LAAIIEE635 pKa = 4.49DD636 pKa = 3.83ALTTDD641 pKa = 3.43RR642 pKa = 11.84SVIQLVVDD650 pKa = 4.45GKK652 pKa = 9.09VLRR655 pKa = 11.84ANVTEE660 pKa = 4.21GDD662 pKa = 3.51TDD664 pKa = 3.59EE665 pKa = 4.4VVANIIEE672 pKa = 4.16QWKK675 pKa = 10.19LDD677 pKa = 3.53SDD679 pKa = 4.32YY680 pKa = 11.29STLSEE685 pKa = 4.0NQMEE689 pKa = 4.46YY690 pKa = 9.21WGDD693 pKa = 3.37YY694 pKa = 10.41ARR696 pKa = 11.84IRR698 pKa = 11.84FDD700 pKa = 3.03NTEE703 pKa = 3.76SHH705 pKa = 6.15EE706 pKa = 4.42VSVSRR711 pKa = 11.84YY712 pKa = 8.64NGPIEE717 pKa = 3.91TDD719 pKa = 3.52FDD721 pKa = 4.34VVGTVQDD728 pKa = 3.42NYY730 pKa = 10.7EE731 pKa = 3.98RR732 pKa = 11.84EE733 pKa = 3.78AAINFEE739 pKa = 4.13GSHH742 pKa = 7.49IEE744 pKa = 4.13VQALGNNFFNNNGSYY759 pKa = 9.32DD760 pKa = 3.44TDD762 pKa = 3.44AALVVFGATGSINGIDD778 pKa = 4.03VIAGGNDD785 pKa = 3.13YY786 pKa = 10.84FGNYY790 pKa = 10.01SFDD793 pKa = 3.89LPNALAIINIDD804 pKa = 3.38GSVSGEE810 pKa = 3.9INVTTLEE817 pKa = 4.17EE818 pKa = 4.41GASKK822 pKa = 10.0SAYY825 pKa = 9.32MLLQATAGEE834 pKa = 4.33LSLDD838 pKa = 3.52DD839 pKa = 3.39VTINVTAAGNNSSADD854 pKa = 3.75LEE856 pKa = 4.56IGGDD860 pKa = 4.07LNNLPIPGFWSTDD873 pKa = 3.25SVNTLVTGSINGITVSASGDD893 pKa = 3.5NSDD896 pKa = 4.4ASLDD900 pKa = 3.43ISGDD904 pKa = 2.92ITIRR908 pKa = 11.84GTILVEE914 pKa = 4.44TSAEE918 pKa = 3.95DD919 pKa = 3.49ANAEE923 pKa = 4.11LSIDD927 pKa = 4.05SNFDD931 pKa = 3.29SSDD934 pKa = 3.54TLSSSVITFDD944 pKa = 3.52NVDD947 pKa = 3.91IIARR951 pKa = 11.84AYY953 pKa = 10.46GEE955 pKa = 4.26SSDD958 pKa = 3.83VDD960 pKa = 3.23ISIDD964 pKa = 3.27GNAYY968 pKa = 10.32GSINSILLVAHH979 pKa = 7.38ADD981 pKa = 3.61GALDD985 pKa = 3.81GSRR988 pKa = 11.84NAEE991 pKa = 4.05SADD994 pKa = 3.62LNVDD998 pKa = 3.08INFNGTVGDD1007 pKa = 3.59ITLQAMGNSDD1017 pKa = 5.66DD1018 pKa = 4.71VDD1020 pKa = 5.32LDD1022 pKa = 3.35MDD1024 pKa = 4.37ASSNRR1029 pKa = 11.84SNGDD1033 pKa = 2.92EE1034 pKa = 4.07VQFYY1038 pKa = 11.19DD1039 pKa = 4.15LGQMMSYY1046 pKa = 10.72DD1047 pKa = 4.32SSWQDD1052 pKa = 3.37LQTGDD1057 pKa = 5.17LISLTIGDD1065 pKa = 4.26VVLTATVGQDD1075 pKa = 3.07VNDD1078 pKa = 4.55DD1079 pKa = 3.52YY1080 pKa = 11.52GTAVEE1085 pKa = 4.31VLADD1089 pKa = 3.81VVSQLQDD1096 pKa = 2.99AVDD1099 pKa = 3.82ARR1101 pKa = 11.84NVDD1104 pKa = 3.7PDD1106 pKa = 4.04LSDD1109 pKa = 3.99LNVVIKK1115 pKa = 8.66QTGNTNYY1122 pKa = 9.92FDD1124 pKa = 3.9NYY1126 pKa = 10.42YY1127 pKa = 10.26GADD1130 pKa = 4.39GIWVRR1135 pKa = 11.84WLDD1138 pKa = 3.67AGEE1141 pKa = 4.17QEE1143 pKa = 4.28ATSLNVDD1150 pKa = 3.5GDD1152 pKa = 4.02IVSPTDD1158 pKa = 3.25RR1159 pKa = 11.84EE1160 pKa = 4.35YY1161 pKa = 11.68GNDD1164 pKa = 3.34MPTSIEE1170 pKa = 3.99FLDD1173 pKa = 3.93SNILIEE1179 pKa = 4.17AHH1181 pKa = 6.46GEE1183 pKa = 4.08SSDD1186 pKa = 4.41ASLDD1190 pKa = 3.74VNDD1193 pKa = 3.4ATGRR1197 pKa = 11.84INSLTLIATDD1207 pKa = 3.98EE1208 pKa = 4.46NGGSADD1214 pKa = 3.13VDD1216 pKa = 3.55ADD1218 pKa = 3.55IEE1220 pKa = 4.4LGGRR1224 pKa = 11.84LGDD1227 pKa = 3.61VTITADD1233 pKa = 3.76GYY1235 pKa = 8.51NTEE1238 pKa = 4.68AQVEE1242 pKa = 4.52MNSAPEE1248 pKa = 3.9GLTYY1252 pKa = 10.91DD1253 pKa = 3.95NSIITLNALGDD1264 pKa = 4.05SADD1267 pKa = 3.66VEE1269 pKa = 4.61YY1270 pKa = 10.69HH1271 pKa = 5.78QEE1273 pKa = 4.08DD1274 pKa = 4.47YY1275 pKa = 11.53GQGTIDD1281 pKa = 3.55SLNINVIGNNYY1292 pKa = 9.78ADD1294 pKa = 4.14ADD1296 pKa = 4.23VIIGGDD1302 pKa = 3.02ITFDD1306 pKa = 4.11SEE1308 pKa = 3.93INLYY1312 pKa = 10.27ARR1314 pKa = 11.84NTEE1317 pKa = 3.87SSDD1320 pKa = 3.21GVNIYY1325 pKa = 8.64MHH1327 pKa = 7.32LANEE1331 pKa = 4.32DD1332 pKa = 3.46RR1333 pKa = 11.84DD1334 pKa = 4.18DD1335 pKa = 4.0GMNSTFHH1342 pKa = 6.44MNDD1345 pKa = 2.41VTINVEE1351 pKa = 4.24SVGYY1355 pKa = 8.95NARR1358 pKa = 11.84AEE1360 pKa = 4.18ITTQNYY1366 pKa = 9.22AHH1368 pKa = 7.12TDD1370 pKa = 3.18GDD1372 pKa = 4.02FSSDD1376 pKa = 3.19SSIDD1380 pKa = 3.39STITGTINALNVSAGVYY1397 pKa = 10.16DD1398 pKa = 5.25DD1399 pKa = 4.58SNEE1402 pKa = 4.49SINPTMVANAEE1413 pKa = 4.2SSVDD1417 pKa = 3.95LNVHH1421 pKa = 6.63GIIEE1425 pKa = 4.99SITLVADD1432 pKa = 3.5NVSEE1436 pKa = 4.23VSVDD1440 pKa = 3.45LFGAINANGVMDD1452 pKa = 3.65ITITNSNSSSISLDD1466 pKa = 3.33LYY1468 pKa = 11.08DD1469 pKa = 3.75VYY1471 pKa = 11.69GDD1473 pKa = 3.83IEE1475 pKa = 5.0TITSNSGSEE1484 pKa = 4.01GAEE1487 pKa = 3.76TNIYY1491 pKa = 10.46VSTEE1495 pKa = 3.68GSIQTVNLTTTQDD1508 pKa = 3.4AEE1510 pKa = 4.65TVASFDD1516 pKa = 3.82INGSQHH1522 pKa = 6.56YY1523 pKa = 8.96INNMTITTVGTSFFGDD1539 pKa = 3.63YY1540 pKa = 10.52TSVSDD1545 pKa = 4.75VIVDD1549 pKa = 3.69QEE1551 pKa = 4.08AHH1553 pKa = 6.09GGVVNIVNGTSNGDD1567 pKa = 3.59LLADD1571 pKa = 3.87TRR1573 pKa = 11.84LSYY1576 pKa = 10.57YY1577 pKa = 11.02GNYY1580 pKa = 9.05QADD1583 pKa = 4.3SIVIAANNNDD1593 pKa = 3.42GAGDD1597 pKa = 3.72EE1598 pKa = 4.43FALHH1602 pKa = 6.81ISMDD1606 pKa = 3.93DD1607 pKa = 3.17VDD1609 pKa = 4.88VYY1611 pKa = 10.63STQHH1615 pKa = 4.36QTALVNNMTSITGLVNGYY1633 pKa = 10.41DD1634 pKa = 3.37NPTNNFIQFADD1645 pKa = 4.65LEE1647 pKa = 5.07DD1648 pKa = 4.95DD1649 pKa = 4.42DD1650 pKa = 4.4YY1651 pKa = 11.66TYY1653 pKa = 11.38EE1654 pKa = 4.38PGANGRR1660 pKa = 11.84QGPIDD1665 pKa = 4.55LNISHH1670 pKa = 6.84YY1671 pKa = 11.49SNGLDD1676 pKa = 3.07GWVGGDD1682 pKa = 3.67TYY1684 pKa = 11.4FDD1686 pKa = 4.83DD1687 pKa = 5.32YY1688 pKa = 11.52EE1689 pKa = 5.04GSQNIGNGFYY1699 pKa = 11.04GSFNEE1704 pKa = 5.12FFDD1707 pKa = 3.82AAQFGIASEE1716 pKa = 4.39YY1717 pKa = 11.32AFVEE1721 pKa = 4.46LSQYY1725 pKa = 9.89GAAGHH1730 pKa = 7.01DD1731 pKa = 4.05GYY1733 pKa = 11.44DD1734 pKa = 3.24FGFDD1738 pKa = 2.84TDD1740 pKa = 3.66VYY1742 pKa = 11.41LNVSWVDD1749 pKa = 3.43KK1750 pKa = 10.91NGNSQTIYY1758 pKa = 11.05LNADD1762 pKa = 3.47LGGSSLGGSLAGDD1775 pKa = 3.65ATAVVNGLNADD1786 pKa = 4.01TDD1788 pKa = 3.72WDD1790 pKa = 4.01GTPTGSLDD1798 pKa = 3.71GSDD1801 pKa = 4.26GRR1803 pKa = 11.84WDD1805 pKa = 3.25SWDD1808 pKa = 3.42NNLTVFANDD1817 pKa = 3.34SNTLGIRR1824 pKa = 11.84IGEE1827 pKa = 4.27GYY1829 pKa = 7.54ITNVSFHH1836 pKa = 6.9SDD1838 pKa = 2.58WVEE1841 pKa = 3.79VDD1843 pKa = 5.01DD1844 pKa = 6.05LVNDD1848 pKa = 3.64YY1849 pKa = 10.9TNIGSSDD1856 pKa = 3.13ASQSKK1861 pKa = 10.62GYY1863 pKa = 9.29TSLNDD1868 pKa = 3.2YY1869 pKa = 11.7ANFEE1873 pKa = 4.13EE1874 pKa = 5.24SSIQFVEE1881 pKa = 4.09GGGYY1885 pKa = 9.81FFGMVLNEE1893 pKa = 4.29DD1894 pKa = 3.57QNSYY1898 pKa = 10.83RR1899 pKa = 11.84GVLAMDD1905 pKa = 3.89EE1906 pKa = 4.54DD1907 pKa = 5.01GEE1909 pKa = 4.82GVTKK1913 pKa = 10.76LIEE1916 pKa = 4.29LVDD1919 pKa = 4.04VYY1921 pKa = 11.56ASYY1924 pKa = 11.9NDD1926 pKa = 2.72VDD1928 pKa = 3.94YY1929 pKa = 11.49YY1930 pKa = 11.55LNEE1933 pKa = 3.96GDD1935 pKa = 5.11ADD1937 pKa = 3.44HH1938 pKa = 7.27GYY1940 pKa = 11.21NDD1942 pKa = 4.15FSSAYY1947 pKa = 9.68IGMGYY1952 pKa = 10.31GNEE1955 pKa = 4.19GGGFAGYY1962 pKa = 10.34AGSTGFDD1969 pKa = 3.66YY1970 pKa = 8.1EE1971 pKa = 4.55TNADD1975 pKa = 3.18NYY1977 pKa = 11.19AFVVV1981 pKa = 3.57
Molecular weight: 209.9 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B7IY23|A0A0B7IY23_9PROT XRE family transcriptional regulator OS=Candidatus Methylopumilus turicensis OX=1581680 GN=BN1209_0365 PE=4 SV=1
MM1 pKa = 7.54ALIKK5 pKa = 10.15VKK7 pKa = 8.97PTSPGRR13 pKa = 11.84RR14 pKa = 11.84AVVKK18 pKa = 10.45VVTPDD23 pKa = 3.35LYY25 pKa = 10.83KK26 pKa = 10.79GKK28 pKa = 9.5PYY30 pKa = 11.01APLLEE35 pKa = 4.52KK36 pKa = 10.46QSKK39 pKa = 8.96NAGRR43 pKa = 11.84NNNGHH48 pKa = 5.15ITIRR52 pKa = 11.84HH53 pKa = 5.01QGGGHH58 pKa = 5.1KK59 pKa = 8.87QHH61 pKa = 6.13YY62 pKa = 9.02RR63 pKa = 11.84LVDD66 pKa = 3.89FRR68 pKa = 11.84RR69 pKa = 11.84NKK71 pKa = 10.58DD72 pKa = 3.41GIPAKK77 pKa = 10.41VEE79 pKa = 4.18HH80 pKa = 6.97IEE82 pKa = 3.9YY83 pKa = 10.62DD84 pKa = 3.75PNRR87 pKa = 11.84SAHH90 pKa = 6.51IALLCYY96 pKa = 10.35ADD98 pKa = 3.41GEE100 pKa = 4.27RR101 pKa = 11.84RR102 pKa = 11.84YY103 pKa = 10.13IIAPRR108 pKa = 11.84GVAAGAQLISGSDD121 pKa = 3.44APIKK125 pKa = 10.53VGNALPLRR133 pKa = 11.84NIPVGSTIHH142 pKa = 6.89CIEE145 pKa = 4.24LQPGKK150 pKa = 10.25GAQLARR156 pKa = 11.84SAGTSVQLLARR167 pKa = 11.84EE168 pKa = 4.17GSYY171 pKa = 10.98AQLRR175 pKa = 11.84LRR177 pKa = 11.84SGEE180 pKa = 3.81VRR182 pKa = 11.84RR183 pKa = 11.84VHH185 pKa = 6.63VDD187 pKa = 3.58CKK189 pKa = 10.37ATLGEE194 pKa = 4.38VGNEE198 pKa = 3.83EE199 pKa = 3.77HH200 pKa = 6.9SLRR203 pKa = 11.84SIGKK207 pKa = 8.64AGAMRR212 pKa = 11.84WRR214 pKa = 11.84GVRR217 pKa = 11.84PTVRR221 pKa = 11.84GVVMNPVDD229 pKa = 3.77HH230 pKa = 6.67PHH232 pKa = 6.77GGGEE236 pKa = 4.36GKK238 pKa = 7.89TAAGMNPVSPWGTPTKK254 pKa = 10.48GYY256 pKa = 7.67RR257 pKa = 11.84TRR259 pKa = 11.84SNKK262 pKa = 8.78RR263 pKa = 11.84TDD265 pKa = 2.85NMRR268 pKa = 11.84VSRR271 pKa = 11.84RR272 pKa = 11.84PANKK276 pKa = 9.73RR277 pKa = 3.2
MM1 pKa = 7.54ALIKK5 pKa = 10.15VKK7 pKa = 8.97PTSPGRR13 pKa = 11.84RR14 pKa = 11.84AVVKK18 pKa = 10.45VVTPDD23 pKa = 3.35LYY25 pKa = 10.83KK26 pKa = 10.79GKK28 pKa = 9.5PYY30 pKa = 11.01APLLEE35 pKa = 4.52KK36 pKa = 10.46QSKK39 pKa = 8.96NAGRR43 pKa = 11.84NNNGHH48 pKa = 5.15ITIRR52 pKa = 11.84HH53 pKa = 5.01QGGGHH58 pKa = 5.1KK59 pKa = 8.87QHH61 pKa = 6.13YY62 pKa = 9.02RR63 pKa = 11.84LVDD66 pKa = 3.89FRR68 pKa = 11.84RR69 pKa = 11.84NKK71 pKa = 10.58DD72 pKa = 3.41GIPAKK77 pKa = 10.41VEE79 pKa = 4.18HH80 pKa = 6.97IEE82 pKa = 3.9YY83 pKa = 10.62DD84 pKa = 3.75PNRR87 pKa = 11.84SAHH90 pKa = 6.51IALLCYY96 pKa = 10.35ADD98 pKa = 3.41GEE100 pKa = 4.27RR101 pKa = 11.84RR102 pKa = 11.84YY103 pKa = 10.13IIAPRR108 pKa = 11.84GVAAGAQLISGSDD121 pKa = 3.44APIKK125 pKa = 10.53VGNALPLRR133 pKa = 11.84NIPVGSTIHH142 pKa = 6.89CIEE145 pKa = 4.24LQPGKK150 pKa = 10.25GAQLARR156 pKa = 11.84SAGTSVQLLARR167 pKa = 11.84EE168 pKa = 4.17GSYY171 pKa = 10.98AQLRR175 pKa = 11.84LRR177 pKa = 11.84SGEE180 pKa = 3.81VRR182 pKa = 11.84RR183 pKa = 11.84VHH185 pKa = 6.63VDD187 pKa = 3.58CKK189 pKa = 10.37ATLGEE194 pKa = 4.38VGNEE198 pKa = 3.83EE199 pKa = 3.77HH200 pKa = 6.9SLRR203 pKa = 11.84SIGKK207 pKa = 8.64AGAMRR212 pKa = 11.84WRR214 pKa = 11.84GVRR217 pKa = 11.84PTVRR221 pKa = 11.84GVVMNPVDD229 pKa = 3.77HH230 pKa = 6.67PHH232 pKa = 6.77GGGEE236 pKa = 4.36GKK238 pKa = 7.89TAAGMNPVSPWGTPTKK254 pKa = 10.48GYY256 pKa = 7.67RR257 pKa = 11.84TRR259 pKa = 11.84SNKK262 pKa = 8.78RR263 pKa = 11.84TDD265 pKa = 2.85NMRR268 pKa = 11.84VSRR271 pKa = 11.84RR272 pKa = 11.84PANKK276 pKa = 9.73RR277 pKa = 3.2
Molecular weight: 30.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
534910 |
37 |
2061 |
307.6 |
33.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.883 ± 0.067 | 0.902 ± 0.019 |
5.362 ± 0.045 | 5.835 ± 0.054 |
4.003 ± 0.046 | 7.043 ± 0.063 |
2.325 ± 0.026 | 6.421 ± 0.045 |
5.73 ± 0.059 | 10.344 ± 0.065 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.766 ± 0.025 | 4.144 ± 0.046 |
4.126 ± 0.032 | 4.138 ± 0.038 |
4.705 ± 0.048 | 6.102 ± 0.055 |
5.2 ± 0.04 | 7.04 ± 0.048 |
1.156 ± 0.022 | 2.776 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
