
Circoviridae 10 LDMD-2013
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; environmental samples
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
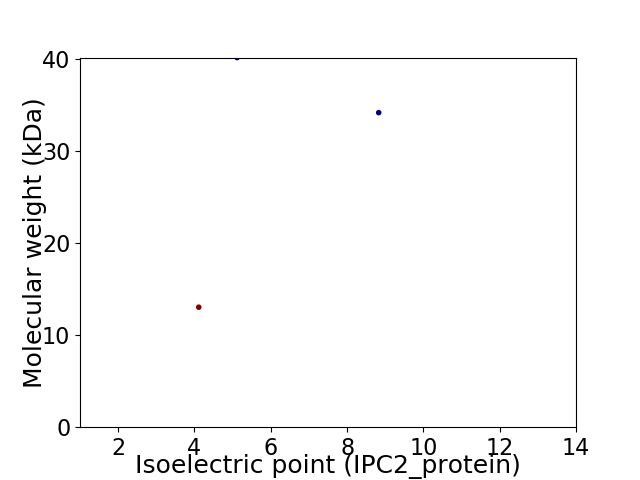
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
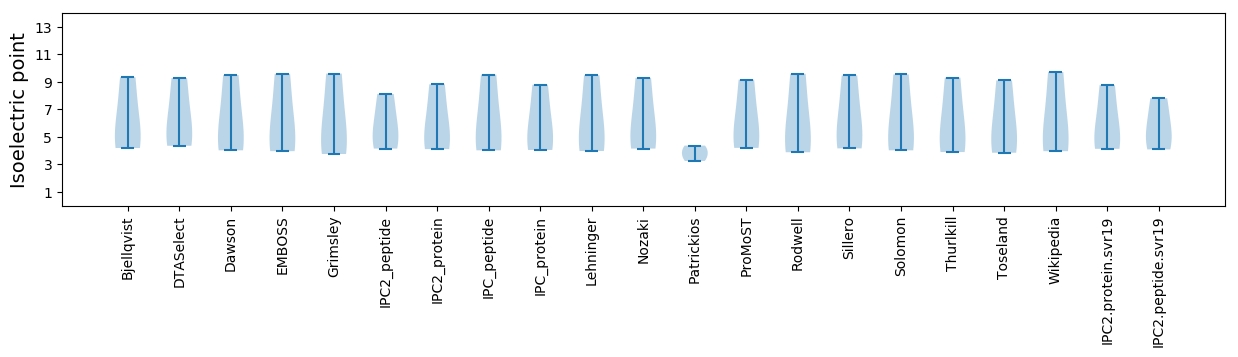
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S5T687|S5T687_9CIRC Uncharacterized protein OS=Circoviridae 10 LDMD-2013 OX=1379714 PE=4 SV=1
MM1 pKa = 8.1DD2 pKa = 5.99PGDD5 pKa = 4.43PDD7 pKa = 3.72FDD9 pKa = 4.91AYY11 pKa = 10.77AGLPIQANVGSPAEE25 pKa = 4.11PADD28 pKa = 3.35PHH30 pKa = 7.19NLVNQGVRR38 pKa = 11.84ALILAGGEE46 pKa = 4.08LVRR49 pKa = 11.84QYY51 pKa = 11.72LFGRR55 pKa = 11.84EE56 pKa = 4.0LLPAEE61 pKa = 4.68RR62 pKa = 11.84LIEE65 pKa = 4.59DD66 pKa = 4.93PPPPPYY72 pKa = 10.57DD73 pKa = 4.03PSDD76 pKa = 3.78SDD78 pKa = 3.95SSSSSSEE85 pKa = 4.1DD86 pKa = 3.19EE87 pKa = 4.35GPYY90 pKa = 10.22PLNTTDD96 pKa = 3.91QIGYY100 pKa = 9.22KK101 pKa = 9.52YY102 pKa = 10.32AGRR105 pKa = 11.84APTVEE110 pKa = 4.02QYY112 pKa = 10.88LARR115 pKa = 11.84NVQKK119 pKa = 9.87TSS121 pKa = 2.82
MM1 pKa = 8.1DD2 pKa = 5.99PGDD5 pKa = 4.43PDD7 pKa = 3.72FDD9 pKa = 4.91AYY11 pKa = 10.77AGLPIQANVGSPAEE25 pKa = 4.11PADD28 pKa = 3.35PHH30 pKa = 7.19NLVNQGVRR38 pKa = 11.84ALILAGGEE46 pKa = 4.08LVRR49 pKa = 11.84QYY51 pKa = 11.72LFGRR55 pKa = 11.84EE56 pKa = 4.0LLPAEE61 pKa = 4.68RR62 pKa = 11.84LIEE65 pKa = 4.59DD66 pKa = 4.93PPPPPYY72 pKa = 10.57DD73 pKa = 4.03PSDD76 pKa = 3.78SDD78 pKa = 3.95SSSSSSEE85 pKa = 4.1DD86 pKa = 3.19EE87 pKa = 4.35GPYY90 pKa = 10.22PLNTTDD96 pKa = 3.91QIGYY100 pKa = 9.22KK101 pKa = 9.52YY102 pKa = 10.32AGRR105 pKa = 11.84APTVEE110 pKa = 4.02QYY112 pKa = 10.88LARR115 pKa = 11.84NVQKK119 pKa = 9.87TSS121 pKa = 2.82
Molecular weight: 13.03 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S5TND6|S5TND6_9CIRC ATP-dependent helicase Rep (Fragment) OS=Circoviridae 10 LDMD-2013 OX=1379714 PE=3 SV=1
KKK2 pKa = 9.92DD3 pKa = 3.5YY4 pKa = 9.32KKK6 pKa = 10.54KK7 pKa = 10.53PGPALGTKKK16 pKa = 9.63YYY18 pKa = 9.56NWIFTSYYY26 pKa = 11.11DD27 pKa = 3.48DDD29 pKa = 3.43NPYYY33 pKa = 8.35KK34 pKa = 10.36FSGSPWAQYYY44 pKa = 9.27ICQAEEE50 pKa = 4.26KK51 pKa = 10.76PTTGRR56 pKa = 11.84VHHH59 pKa = 5.01QGYYY63 pKa = 6.08HHH65 pKa = 6.98NTPVGLATLQRR76 pKa = 11.84EEE78 pKa = 4.13GLPGVHHH85 pKa = 6.55EEE87 pKa = 4.12RR88 pKa = 11.84RR89 pKa = 11.84GNHHH93 pKa = 5.35KK94 pKa = 9.09EE95 pKa = 3.27RR96 pKa = 11.84QYYY99 pKa = 10.42RR100 pKa = 11.84KKK102 pKa = 9.9EE103 pKa = 3.93EE104 pKa = 4.03RR105 pKa = 11.84VPGSLPIEEE114 pKa = 4.67GDDD117 pKa = 3.95PQQGSRR123 pKa = 11.84TDDD126 pKa = 4.3DDD128 pKa = 3.73DD129 pKa = 4.61KKK131 pKa = 10.89ILVEEE136 pKa = 4.33GLLGAFEEE144 pKa = 5.0DDD146 pKa = 3.56SSTVRR151 pKa = 11.84FYYY154 pKa = 11.21GFEEE158 pKa = 4.09YYY160 pKa = 9.26SLVGQPRR167 pKa = 11.84TNDDD171 pKa = 3.34AQEEE175 pKa = 3.86YYY177 pKa = 10.59YY178 pKa = 10.36YYY180 pKa = 10.96PPGTGKKK187 pKa = 7.98RR188 pKa = 11.84AVWEEE193 pKa = 4.44VPDDD197 pKa = 4.0DDD199 pKa = 5.45VYYY202 pKa = 11.24CPTSSGKKK210 pKa = 9.43WFDDD214 pKa = 3.54YYY216 pKa = 11.18PGYYY220 pKa = 8.05HH221 pKa = 5.29VILVDDD227 pKa = 4.56DD228 pKa = 4.81YY229 pKa = 11.3GQWPVPFFLQFLDDD243 pKa = 3.84YYY245 pKa = 10.6KK246 pKa = 11.13LLPVKKK252 pKa = 9.95SHHH255 pKa = 5.24HHH257 pKa = 5.57GRR259 pKa = 11.84AKKK262 pKa = 10.14YYY264 pKa = 7.8TSNEEE269 pKa = 4.33IEEE272 pKa = 4.62IYYY275 pKa = 11.26DDD277 pKa = 3.68PAVTIAAIRR286 pKa = 11.84RR287 pKa = 11.84RR288 pKa = 11.84FKKK291 pKa = 10.41EE292 pKa = 3.6KKK294 pKa = 10.23HH295 pKa = 5.91NTL
KKK2 pKa = 9.92DD3 pKa = 3.5YY4 pKa = 9.32KKK6 pKa = 10.54KK7 pKa = 10.53PGPALGTKKK16 pKa = 9.63YYY18 pKa = 9.56NWIFTSYYY26 pKa = 11.11DD27 pKa = 3.48DDD29 pKa = 3.43NPYYY33 pKa = 8.35KK34 pKa = 10.36FSGSPWAQYYY44 pKa = 9.27ICQAEEE50 pKa = 4.26KK51 pKa = 10.76PTTGRR56 pKa = 11.84VHHH59 pKa = 5.01QGYYY63 pKa = 6.08HHH65 pKa = 6.98NTPVGLATLQRR76 pKa = 11.84EEE78 pKa = 4.13GLPGVHHH85 pKa = 6.55EEE87 pKa = 4.12RR88 pKa = 11.84RR89 pKa = 11.84GNHHH93 pKa = 5.35KK94 pKa = 9.09EE95 pKa = 3.27RR96 pKa = 11.84QYYY99 pKa = 10.42RR100 pKa = 11.84KKK102 pKa = 9.9EE103 pKa = 3.93EE104 pKa = 4.03RR105 pKa = 11.84VPGSLPIEEE114 pKa = 4.67GDDD117 pKa = 3.95PQQGSRR123 pKa = 11.84TDDD126 pKa = 4.3DDD128 pKa = 3.73DD129 pKa = 4.61KKK131 pKa = 10.89ILVEEE136 pKa = 4.33GLLGAFEEE144 pKa = 5.0DDD146 pKa = 3.56SSTVRR151 pKa = 11.84FYYY154 pKa = 11.21GFEEE158 pKa = 4.09YYY160 pKa = 9.26SLVGQPRR167 pKa = 11.84TNDDD171 pKa = 3.34AQEEE175 pKa = 3.86YYY177 pKa = 10.59YY178 pKa = 10.36YYY180 pKa = 10.96PPGTGKKK187 pKa = 7.98RR188 pKa = 11.84AVWEEE193 pKa = 4.44VPDDD197 pKa = 4.0DDD199 pKa = 5.45VYYY202 pKa = 11.24CPTSSGKKK210 pKa = 9.43WFDDD214 pKa = 3.54YYY216 pKa = 11.18PGYYY220 pKa = 8.05HH221 pKa = 5.29VILVDDD227 pKa = 4.56DD228 pKa = 4.81YY229 pKa = 11.3GQWPVPFFLQFLDDD243 pKa = 3.84YYY245 pKa = 10.6KK246 pKa = 11.13LLPVKKK252 pKa = 9.95SHHH255 pKa = 5.24HHH257 pKa = 5.57GRR259 pKa = 11.84AKKK262 pKa = 10.14YYY264 pKa = 7.8TSNEEE269 pKa = 4.33IEEE272 pKa = 4.62IYYY275 pKa = 11.26DDD277 pKa = 3.68PAVTIAAIRR286 pKa = 11.84RR287 pKa = 11.84RR288 pKa = 11.84FKKK291 pKa = 10.41EE292 pKa = 3.6KKK294 pKa = 10.23HH295 pKa = 5.91NTL
Molecular weight: 34.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
786 |
121 |
367 |
262.0 |
29.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.489 ± 0.835 | 0.763 ± 0.308 |
6.489 ± 0.835 | 5.344 ± 0.388 |
4.326 ± 0.875 | 8.906 ± 0.26 |
2.163 ± 0.422 | 4.326 ± 0.33 |
4.962 ± 1.033 | 6.616 ± 0.92 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.272 ± 0.547 | 4.326 ± 1.029 |
8.142 ± 1.727 | 3.308 ± 0.76 |
5.344 ± 1.503 | 7.506 ± 1.352 |
5.98 ± 0.829 | 6.489 ± 0.529 |
1.527 ± 0.616 | 5.598 ± 0.422 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
