
Beihai picorna-like virus 117
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.39
Get precalculated fractions of proteins
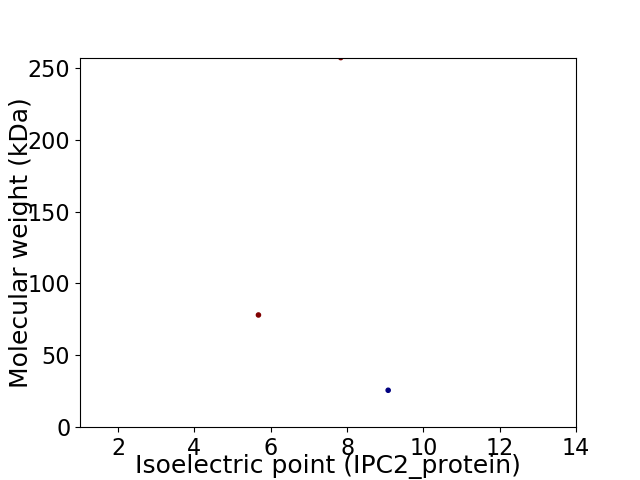
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KHE6|A0A1L3KHE6_9VIRU Uncharacterized protein OS=Beihai picorna-like virus 117 OX=1922546 PE=4 SV=1
MM1 pKa = 8.18ASGQPLATAAPVVDD15 pKa = 4.8SAATPAGPLSLEE27 pKa = 4.36TGRR30 pKa = 11.84TTHH33 pKa = 6.08QPLSTVTGNFNSLDD47 pKa = 3.55PFLYY51 pKa = 9.81RR52 pKa = 11.84HH53 pKa = 5.94NFIIANFRR61 pKa = 11.84WNADD65 pKa = 3.39KK66 pKa = 11.34APGNLIWQLPLSPYY80 pKa = 10.61NLGGRR85 pKa = 11.84IAEE88 pKa = 3.98RR89 pKa = 11.84AAAYY93 pKa = 9.1LYY95 pKa = 10.09WRR97 pKa = 11.84GQISFEE103 pKa = 4.42VIVVATGFHH112 pKa = 6.71AGQLFIVEE120 pKa = 4.7TPPDD124 pKa = 4.09YY125 pKa = 11.11KK126 pKa = 10.66PGSTQSISDD135 pKa = 3.98LSTFHH140 pKa = 7.19HH141 pKa = 6.22EE142 pKa = 4.21QVDD145 pKa = 4.05VKK147 pKa = 11.13VGVPIQFSTRR157 pKa = 11.84DD158 pKa = 3.11ICPTQFHH165 pKa = 5.97TVFDD169 pKa = 4.11SQSTLKK175 pKa = 9.24PTEE178 pKa = 4.42SVTGMQSIGGYY189 pKa = 9.78ISMFNLTKK197 pKa = 10.01LTAANTAPEE206 pKa = 3.49ISIIVKK212 pKa = 10.36ACLDD216 pKa = 3.54PSFEE220 pKa = 4.13VSIMRR225 pKa = 11.84PLSFDD230 pKa = 3.39NEE232 pKa = 4.01RR233 pKa = 11.84DD234 pKa = 3.55EE235 pKa = 5.23VEE237 pKa = 3.91TGRR240 pKa = 11.84WQLLAEE246 pKa = 4.45RR247 pKa = 11.84LNDD250 pKa = 3.38NAKK253 pKa = 10.73DD254 pKa = 3.9YY255 pKa = 11.03GVKK258 pKa = 10.64DD259 pKa = 3.4SDD261 pKa = 3.86VAGLFLVGGSTEE273 pKa = 3.39AWRR276 pKa = 11.84GVDD279 pKa = 3.74RR280 pKa = 11.84LGNAANPYY288 pKa = 8.73TGRR291 pKa = 11.84NMEE294 pKa = 4.66KK295 pKa = 10.69LLMAPCSITGNVNKK309 pKa = 10.29EE310 pKa = 4.11DD311 pKa = 3.94IYY313 pKa = 11.45LSDD316 pKa = 5.15DD317 pKa = 3.59YY318 pKa = 11.69TPITHH323 pKa = 6.71PVGRR327 pKa = 11.84FNMKK331 pKa = 9.93IVDD334 pKa = 3.73PEE336 pKa = 4.02FNAVKK341 pKa = 10.05NQTYY345 pKa = 8.95FYY347 pKa = 8.8PIAYY351 pKa = 6.49GTSGSATYY359 pKa = 9.31TKK361 pKa = 9.74YY362 pKa = 9.54TNTDD366 pKa = 2.6WYY368 pKa = 10.07LGRR371 pKa = 11.84VSSDD375 pKa = 2.48KK376 pKa = 11.34YY377 pKa = 10.0YY378 pKa = 11.16VCTNPTRR385 pKa = 11.84TSFHH389 pKa = 6.35KK390 pKa = 10.85VITPSGYY397 pKa = 9.78CLHH400 pKa = 6.57VKK402 pKa = 10.2PSDD405 pKa = 3.18ITGPSVTALNGEE417 pKa = 3.99NLLYY421 pKa = 9.85TRR423 pKa = 11.84CGTGSVAGTLTEE435 pKa = 4.32SDD437 pKa = 3.68SLCFKK442 pKa = 10.6EE443 pKa = 3.96ITEE446 pKa = 4.25FCIQHH451 pKa = 7.0KK452 pKa = 8.77ITLTSNTPVFRR463 pKa = 11.84ILDD466 pKa = 3.72RR467 pKa = 11.84VTKK470 pKa = 10.27QVLGHH475 pKa = 6.59ARR477 pKa = 11.84LNKK480 pKa = 10.01QGFLSTNFPSEE491 pKa = 3.99TVVFYY496 pKa = 10.9EE497 pKa = 4.18DD498 pKa = 2.98VMLAFEE504 pKa = 4.56GMYY507 pKa = 10.3SEE509 pKa = 6.34LEE511 pKa = 4.1DD512 pKa = 3.98LPEE515 pKa = 5.37ISVTASTYY523 pKa = 10.92LLQKK527 pKa = 11.03AMTRR531 pKa = 11.84SFEE534 pKa = 4.46RR535 pKa = 11.84MVNKK539 pKa = 10.46NKK541 pKa = 9.65TKK543 pKa = 10.25SKK545 pKa = 10.85NGISSSSYY553 pKa = 8.16GSSRR557 pKa = 11.84RR558 pKa = 11.84FRR560 pKa = 11.84GRR562 pKa = 11.84RR563 pKa = 11.84APVVYY568 pKa = 10.07HH569 pKa = 5.77RR570 pKa = 11.84QSYY573 pKa = 10.93DD574 pKa = 3.02GGKK577 pKa = 9.91EE578 pKa = 3.65EE579 pKa = 4.01PTYY582 pKa = 11.04DD583 pKa = 3.47RR584 pKa = 11.84EE585 pKa = 4.4SSQEE589 pKa = 3.75RR590 pKa = 11.84YY591 pKa = 8.51EE592 pKa = 4.32SKK594 pKa = 10.16RR595 pKa = 11.84SQRR598 pKa = 11.84RR599 pKa = 11.84FKK601 pKa = 10.95RR602 pKa = 11.84KK603 pKa = 8.05TNTLHH608 pKa = 6.67SKK610 pKa = 10.4RR611 pKa = 11.84SEE613 pKa = 4.06ATDD616 pKa = 4.0DD617 pKa = 4.2GSDD620 pKa = 3.6SEE622 pKa = 5.22FEE624 pKa = 3.64ILEE627 pKa = 4.13YY628 pKa = 10.3PEE630 pKa = 4.67CSNPEE635 pKa = 3.67RR636 pKa = 11.84TEE638 pKa = 4.17CPEE641 pKa = 4.22CPAPGWPHH649 pKa = 7.01ADD651 pKa = 3.14VDD653 pKa = 4.3PEE655 pKa = 4.39SGEE658 pKa = 3.95LTDD661 pKa = 4.99VSNEE665 pKa = 3.82PTGEE669 pKa = 4.04RR670 pKa = 11.84RR671 pKa = 11.84DD672 pKa = 3.76SSIDD676 pKa = 3.32ALLEE680 pKa = 3.96RR681 pKa = 11.84LTQLGARR688 pKa = 11.84ATRR691 pKa = 11.84VFGRR695 pKa = 11.84RR696 pKa = 11.84NLQDD700 pKa = 3.07VV701 pKa = 3.66
MM1 pKa = 8.18ASGQPLATAAPVVDD15 pKa = 4.8SAATPAGPLSLEE27 pKa = 4.36TGRR30 pKa = 11.84TTHH33 pKa = 6.08QPLSTVTGNFNSLDD47 pKa = 3.55PFLYY51 pKa = 9.81RR52 pKa = 11.84HH53 pKa = 5.94NFIIANFRR61 pKa = 11.84WNADD65 pKa = 3.39KK66 pKa = 11.34APGNLIWQLPLSPYY80 pKa = 10.61NLGGRR85 pKa = 11.84IAEE88 pKa = 3.98RR89 pKa = 11.84AAAYY93 pKa = 9.1LYY95 pKa = 10.09WRR97 pKa = 11.84GQISFEE103 pKa = 4.42VIVVATGFHH112 pKa = 6.71AGQLFIVEE120 pKa = 4.7TPPDD124 pKa = 4.09YY125 pKa = 11.11KK126 pKa = 10.66PGSTQSISDD135 pKa = 3.98LSTFHH140 pKa = 7.19HH141 pKa = 6.22EE142 pKa = 4.21QVDD145 pKa = 4.05VKK147 pKa = 11.13VGVPIQFSTRR157 pKa = 11.84DD158 pKa = 3.11ICPTQFHH165 pKa = 5.97TVFDD169 pKa = 4.11SQSTLKK175 pKa = 9.24PTEE178 pKa = 4.42SVTGMQSIGGYY189 pKa = 9.78ISMFNLTKK197 pKa = 10.01LTAANTAPEE206 pKa = 3.49ISIIVKK212 pKa = 10.36ACLDD216 pKa = 3.54PSFEE220 pKa = 4.13VSIMRR225 pKa = 11.84PLSFDD230 pKa = 3.39NEE232 pKa = 4.01RR233 pKa = 11.84DD234 pKa = 3.55EE235 pKa = 5.23VEE237 pKa = 3.91TGRR240 pKa = 11.84WQLLAEE246 pKa = 4.45RR247 pKa = 11.84LNDD250 pKa = 3.38NAKK253 pKa = 10.73DD254 pKa = 3.9YY255 pKa = 11.03GVKK258 pKa = 10.64DD259 pKa = 3.4SDD261 pKa = 3.86VAGLFLVGGSTEE273 pKa = 3.39AWRR276 pKa = 11.84GVDD279 pKa = 3.74RR280 pKa = 11.84LGNAANPYY288 pKa = 8.73TGRR291 pKa = 11.84NMEE294 pKa = 4.66KK295 pKa = 10.69LLMAPCSITGNVNKK309 pKa = 10.29EE310 pKa = 4.11DD311 pKa = 3.94IYY313 pKa = 11.45LSDD316 pKa = 5.15DD317 pKa = 3.59YY318 pKa = 11.69TPITHH323 pKa = 6.71PVGRR327 pKa = 11.84FNMKK331 pKa = 9.93IVDD334 pKa = 3.73PEE336 pKa = 4.02FNAVKK341 pKa = 10.05NQTYY345 pKa = 8.95FYY347 pKa = 8.8PIAYY351 pKa = 6.49GTSGSATYY359 pKa = 9.31TKK361 pKa = 9.74YY362 pKa = 9.54TNTDD366 pKa = 2.6WYY368 pKa = 10.07LGRR371 pKa = 11.84VSSDD375 pKa = 2.48KK376 pKa = 11.34YY377 pKa = 10.0YY378 pKa = 11.16VCTNPTRR385 pKa = 11.84TSFHH389 pKa = 6.35KK390 pKa = 10.85VITPSGYY397 pKa = 9.78CLHH400 pKa = 6.57VKK402 pKa = 10.2PSDD405 pKa = 3.18ITGPSVTALNGEE417 pKa = 3.99NLLYY421 pKa = 9.85TRR423 pKa = 11.84CGTGSVAGTLTEE435 pKa = 4.32SDD437 pKa = 3.68SLCFKK442 pKa = 10.6EE443 pKa = 3.96ITEE446 pKa = 4.25FCIQHH451 pKa = 7.0KK452 pKa = 8.77ITLTSNTPVFRR463 pKa = 11.84ILDD466 pKa = 3.72RR467 pKa = 11.84VTKK470 pKa = 10.27QVLGHH475 pKa = 6.59ARR477 pKa = 11.84LNKK480 pKa = 10.01QGFLSTNFPSEE491 pKa = 3.99TVVFYY496 pKa = 10.9EE497 pKa = 4.18DD498 pKa = 2.98VMLAFEE504 pKa = 4.56GMYY507 pKa = 10.3SEE509 pKa = 6.34LEE511 pKa = 4.1DD512 pKa = 3.98LPEE515 pKa = 5.37ISVTASTYY523 pKa = 10.92LLQKK527 pKa = 11.03AMTRR531 pKa = 11.84SFEE534 pKa = 4.46RR535 pKa = 11.84MVNKK539 pKa = 10.46NKK541 pKa = 9.65TKK543 pKa = 10.25SKK545 pKa = 10.85NGISSSSYY553 pKa = 8.16GSSRR557 pKa = 11.84RR558 pKa = 11.84FRR560 pKa = 11.84GRR562 pKa = 11.84RR563 pKa = 11.84APVVYY568 pKa = 10.07HH569 pKa = 5.77RR570 pKa = 11.84QSYY573 pKa = 10.93DD574 pKa = 3.02GGKK577 pKa = 9.91EE578 pKa = 3.65EE579 pKa = 4.01PTYY582 pKa = 11.04DD583 pKa = 3.47RR584 pKa = 11.84EE585 pKa = 4.4SSQEE589 pKa = 3.75RR590 pKa = 11.84YY591 pKa = 8.51EE592 pKa = 4.32SKK594 pKa = 10.16RR595 pKa = 11.84SQRR598 pKa = 11.84RR599 pKa = 11.84FKK601 pKa = 10.95RR602 pKa = 11.84KK603 pKa = 8.05TNTLHH608 pKa = 6.67SKK610 pKa = 10.4RR611 pKa = 11.84SEE613 pKa = 4.06ATDD616 pKa = 4.0DD617 pKa = 4.2GSDD620 pKa = 3.6SEE622 pKa = 5.22FEE624 pKa = 3.64ILEE627 pKa = 4.13YY628 pKa = 10.3PEE630 pKa = 4.67CSNPEE635 pKa = 3.67RR636 pKa = 11.84TEE638 pKa = 4.17CPEE641 pKa = 4.22CPAPGWPHH649 pKa = 7.01ADD651 pKa = 3.14VDD653 pKa = 4.3PEE655 pKa = 4.39SGEE658 pKa = 3.95LTDD661 pKa = 4.99VSNEE665 pKa = 3.82PTGEE669 pKa = 4.04RR670 pKa = 11.84RR671 pKa = 11.84DD672 pKa = 3.76SSIDD676 pKa = 3.32ALLEE680 pKa = 3.96RR681 pKa = 11.84LTQLGARR688 pKa = 11.84ATRR691 pKa = 11.84VFGRR695 pKa = 11.84RR696 pKa = 11.84NLQDD700 pKa = 3.07VV701 pKa = 3.66
Molecular weight: 78.09 kDa
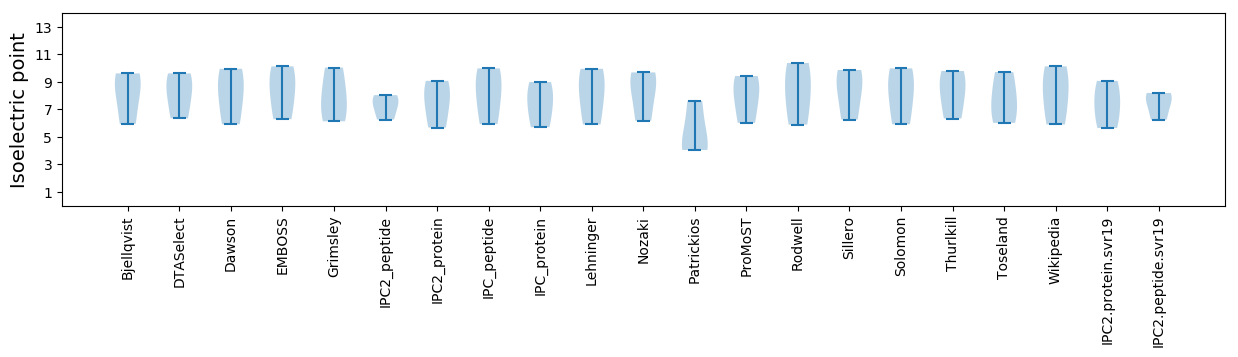
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KHQ4|A0A1L3KHQ4_9VIRU Uncharacterized protein OS=Beihai picorna-like virus 117 OX=1922546 PE=4 SV=1
MM1 pKa = 7.52ASAAALMGAAGVSAGGGLLSSIIDD25 pKa = 3.82NPMMEE30 pKa = 4.19AKK32 pKa = 10.28KK33 pKa = 10.56NRR35 pKa = 11.84LTIEE39 pKa = 4.06NQAKK43 pKa = 10.7NDD45 pKa = 3.78MKK47 pKa = 11.37VNAANEE53 pKa = 4.14GLSEE57 pKa = 4.24KK58 pKa = 9.32LTHH61 pKa = 5.35YY62 pKa = 7.63TANEE66 pKa = 3.83AKK68 pKa = 10.26QLMTAQTANSKK79 pKa = 10.36SLNTQNAQIQKK90 pKa = 9.74EE91 pKa = 4.23LNAQSAQLQAGLMQTSTQNQEE112 pKa = 3.68NLQTFQMNQLEE123 pKa = 4.35KK124 pKa = 11.02DD125 pKa = 4.02GIPRR129 pKa = 11.84SMLYY133 pKa = 10.66LSGSRR138 pKa = 11.84NSVPEE143 pKa = 3.45QRR145 pKa = 11.84AYY147 pKa = 11.13LGGGTFKK154 pKa = 10.56TFNGLSGNTAYY165 pKa = 9.75TGTQAQQMLNMGNPLAFRR183 pKa = 11.84YY184 pKa = 9.4QSRR187 pKa = 11.84NSNFGTRR194 pKa = 11.84TNTASLQKK202 pKa = 10.82GFDD205 pKa = 3.4KK206 pKa = 10.89ASQYY210 pKa = 10.59IGQFKK215 pKa = 9.78PNKK218 pKa = 9.81SKK220 pKa = 11.05GNQVAAYY227 pKa = 8.34SAKK230 pKa = 10.2YY231 pKa = 9.67DD232 pKa = 3.92DD233 pKa = 4.66VMLVPP238 pKa = 5.46
MM1 pKa = 7.52ASAAALMGAAGVSAGGGLLSSIIDD25 pKa = 3.82NPMMEE30 pKa = 4.19AKK32 pKa = 10.28KK33 pKa = 10.56NRR35 pKa = 11.84LTIEE39 pKa = 4.06NQAKK43 pKa = 10.7NDD45 pKa = 3.78MKK47 pKa = 11.37VNAANEE53 pKa = 4.14GLSEE57 pKa = 4.24KK58 pKa = 9.32LTHH61 pKa = 5.35YY62 pKa = 7.63TANEE66 pKa = 3.83AKK68 pKa = 10.26QLMTAQTANSKK79 pKa = 10.36SLNTQNAQIQKK90 pKa = 9.74EE91 pKa = 4.23LNAQSAQLQAGLMQTSTQNQEE112 pKa = 3.68NLQTFQMNQLEE123 pKa = 4.35KK124 pKa = 11.02DD125 pKa = 4.02GIPRR129 pKa = 11.84SMLYY133 pKa = 10.66LSGSRR138 pKa = 11.84NSVPEE143 pKa = 3.45QRR145 pKa = 11.84AYY147 pKa = 11.13LGGGTFKK154 pKa = 10.56TFNGLSGNTAYY165 pKa = 9.75TGTQAQQMLNMGNPLAFRR183 pKa = 11.84YY184 pKa = 9.4QSRR187 pKa = 11.84NSNFGTRR194 pKa = 11.84TNTASLQKK202 pKa = 10.82GFDD205 pKa = 3.4KK206 pKa = 10.89ASQYY210 pKa = 10.59IGQFKK215 pKa = 9.78PNKK218 pKa = 9.81SKK220 pKa = 11.05GNQVAAYY227 pKa = 8.34SAKK230 pKa = 10.2YY231 pKa = 9.67DD232 pKa = 3.92DD233 pKa = 4.66VMLVPP238 pKa = 5.46
Molecular weight: 25.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3218 |
238 |
2279 |
1072.7 |
120.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.433 ± 0.971 | 1.336 ± 0.231 |
5.221 ± 0.486 | 5.5 ± 0.365 |
4.071 ± 0.2 | 5.718 ± 0.796 |
2.859 ± 0.673 | 5.904 ± 1.021 |
6.37 ± 0.774 | 7.955 ± 0.202 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.268 ± 0.48 | 5.252 ± 0.865 |
5.065 ± 0.449 | 4.817 ± 0.934 |
5.003 ± 0.479 | 7.427 ± 0.749 |
8.359 ± 0.227 | 6.091 ± 0.652 |
0.622 ± 0.143 | 3.729 ± 0.141 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
