
McMurdo Ice Shelf pond-associated circular DNA virus-4
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.58
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
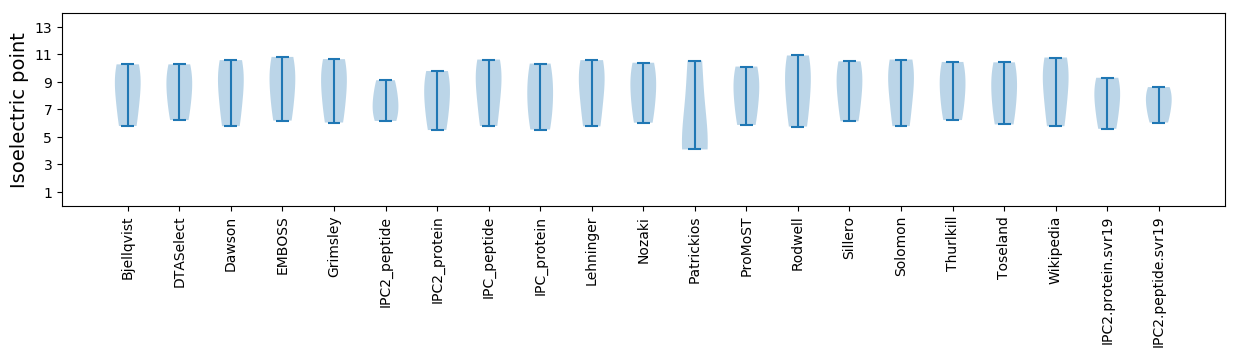
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A075LYS2|A0A075LYS2_9VIRU Replication-associated protein OS=McMurdo Ice Shelf pond-associated circular DNA virus-4 OX=1521388 PE=4 SV=1
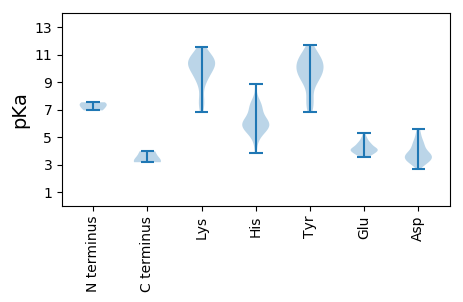
MM1 pKa = 7.3SACEE5 pKa = 3.81TGGVLVHH12 pKa = 6.94AEE14 pKa = 4.12GLASVHH20 pKa = 6.2SPGVVDD26 pKa = 4.85FDD28 pKa = 3.99VLFVCDD34 pKa = 3.1VRR36 pKa = 11.84TRR38 pKa = 11.84VAEE41 pKa = 3.97LKK43 pKa = 10.45VRR45 pKa = 11.84SEE47 pKa = 4.16RR48 pKa = 11.84LVVSEE53 pKa = 4.55DD54 pKa = 3.43TSDD57 pKa = 3.48DD58 pKa = 3.15KK59 pKa = 11.4HH60 pKa = 7.17RR61 pKa = 11.84KK62 pKa = 8.83VLTSVEE68 pKa = 3.98FLQVCIRR75 pKa = 11.84QVRR78 pKa = 11.84VVVVFHH84 pKa = 7.59DD85 pKa = 3.83SGLGKK90 pKa = 10.26LVEE93 pKa = 4.08QLEE96 pKa = 4.38RR97 pKa = 11.84RR98 pKa = 11.84SWFHH102 pKa = 5.47VVRR105 pKa = 11.84LHH107 pKa = 7.45LGDD110 pKa = 5.62LEE112 pKa = 4.54VLDD115 pKa = 4.59KK116 pKa = 11.52LDD118 pKa = 3.58VLEE121 pKa = 4.1QTVRR125 pKa = 11.84LGDD128 pKa = 3.74SAGGTLYY135 pKa = 11.06LVDD138 pKa = 4.42SVHH141 pKa = 7.2SSVTEE146 pKa = 3.97TVVVSQKK153 pKa = 10.76AALNNRR159 pKa = 11.84LEE161 pKa = 4.45CAEE164 pKa = 3.98GRR166 pKa = 11.84TDD168 pKa = 3.27LHH170 pKa = 7.5RR171 pKa = 11.84GDD173 pKa = 4.39VLADD177 pKa = 3.39SAAVVYY183 pKa = 9.96HH184 pKa = 6.31ISSMVRR190 pKa = 11.84HH191 pKa = 5.82VGYY194 pKa = 8.57LTGNLVRR201 pKa = 11.84EE202 pKa = 4.26AGIPRR207 pKa = 11.84CLGSIDD213 pKa = 3.5VVGDD217 pKa = 3.1VRR219 pKa = 11.84EE220 pKa = 3.99NVGIVRR226 pKa = 11.84RR227 pKa = 11.84HH228 pKa = 3.82QQGRR232 pKa = 11.84ARR234 pKa = 11.84CLTVGVVLAVPVAHH248 pKa = 6.2QCPCGGAVDD257 pKa = 5.35LIEE260 pKa = 5.1IEE262 pKa = 4.46EE263 pKa = 4.14VHH265 pKa = 5.9TRR267 pKa = 11.84CLVRR271 pKa = 11.84IDD273 pKa = 3.89GLVLSSYY280 pKa = 10.82DD281 pKa = 3.44VQNALSEE288 pKa = 4.51SSASSGGSSLVGAGGALAGAEE309 pKa = 4.19LSCRR313 pKa = 11.84SSTT316 pKa = 3.97
MM1 pKa = 7.3SACEE5 pKa = 3.81TGGVLVHH12 pKa = 6.94AEE14 pKa = 4.12GLASVHH20 pKa = 6.2SPGVVDD26 pKa = 4.85FDD28 pKa = 3.99VLFVCDD34 pKa = 3.1VRR36 pKa = 11.84TRR38 pKa = 11.84VAEE41 pKa = 3.97LKK43 pKa = 10.45VRR45 pKa = 11.84SEE47 pKa = 4.16RR48 pKa = 11.84LVVSEE53 pKa = 4.55DD54 pKa = 3.43TSDD57 pKa = 3.48DD58 pKa = 3.15KK59 pKa = 11.4HH60 pKa = 7.17RR61 pKa = 11.84KK62 pKa = 8.83VLTSVEE68 pKa = 3.98FLQVCIRR75 pKa = 11.84QVRR78 pKa = 11.84VVVVFHH84 pKa = 7.59DD85 pKa = 3.83SGLGKK90 pKa = 10.26LVEE93 pKa = 4.08QLEE96 pKa = 4.38RR97 pKa = 11.84RR98 pKa = 11.84SWFHH102 pKa = 5.47VVRR105 pKa = 11.84LHH107 pKa = 7.45LGDD110 pKa = 5.62LEE112 pKa = 4.54VLDD115 pKa = 4.59KK116 pKa = 11.52LDD118 pKa = 3.58VLEE121 pKa = 4.1QTVRR125 pKa = 11.84LGDD128 pKa = 3.74SAGGTLYY135 pKa = 11.06LVDD138 pKa = 4.42SVHH141 pKa = 7.2SSVTEE146 pKa = 3.97TVVVSQKK153 pKa = 10.76AALNNRR159 pKa = 11.84LEE161 pKa = 4.45CAEE164 pKa = 3.98GRR166 pKa = 11.84TDD168 pKa = 3.27LHH170 pKa = 7.5RR171 pKa = 11.84GDD173 pKa = 4.39VLADD177 pKa = 3.39SAAVVYY183 pKa = 9.96HH184 pKa = 6.31ISSMVRR190 pKa = 11.84HH191 pKa = 5.82VGYY194 pKa = 8.57LTGNLVRR201 pKa = 11.84EE202 pKa = 4.26AGIPRR207 pKa = 11.84CLGSIDD213 pKa = 3.5VVGDD217 pKa = 3.1VRR219 pKa = 11.84EE220 pKa = 3.99NVGIVRR226 pKa = 11.84RR227 pKa = 11.84HH228 pKa = 3.82QQGRR232 pKa = 11.84ARR234 pKa = 11.84CLTVGVVLAVPVAHH248 pKa = 6.2QCPCGGAVDD257 pKa = 5.35LIEE260 pKa = 5.1IEE262 pKa = 4.46EE263 pKa = 4.14VHH265 pKa = 5.9TRR267 pKa = 11.84CLVRR271 pKa = 11.84IDD273 pKa = 3.89GLVLSSYY280 pKa = 10.82DD281 pKa = 3.44VQNALSEE288 pKa = 4.51SSASSGGSSLVGAGGALAGAEE309 pKa = 4.19LSCRR313 pKa = 11.84SSTT316 pKa = 3.97
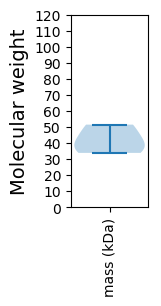
Molecular weight: 33.82 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A075LZ76|A0A075LZ76_9VIRU Uncharacterized protein OS=McMurdo Ice Shelf pond-associated circular DNA virus-4 OX=1521388 PE=4 SV=1
MM1 pKa = 6.94YY2 pKa = 10.17RR3 pKa = 11.84PRR5 pKa = 11.84RR6 pKa = 11.84TTNVGPTGSNPGLAIQRR23 pKa = 11.84LVFGSRR29 pKa = 11.84ARR31 pKa = 11.84EE32 pKa = 3.73EE33 pKa = 4.33DD34 pKa = 3.7SPAVSQGSQAKK45 pKa = 9.34KK46 pKa = 9.78RR47 pKa = 11.84RR48 pKa = 11.84LSRR51 pKa = 11.84SGGWAKK57 pKa = 10.53EE58 pKa = 3.6NRR60 pKa = 11.84TRR62 pKa = 11.84IPLYY66 pKa = 10.28SSDD69 pKa = 3.59MAKK72 pKa = 10.54SSIKK76 pKa = 10.3RR77 pKa = 11.84SGSRR81 pKa = 11.84IAKK84 pKa = 9.25RR85 pKa = 11.84RR86 pKa = 11.84YY87 pKa = 7.09TKK89 pKa = 10.45RR90 pKa = 11.84STKK93 pKa = 10.01GRR95 pKa = 11.84TRR97 pKa = 11.84RR98 pKa = 11.84PLSRR102 pKa = 11.84ARR104 pKa = 11.84PTRR107 pKa = 11.84KK108 pKa = 8.94FRR110 pKa = 11.84TRR112 pKa = 11.84KK113 pKa = 9.9SPARR117 pKa = 11.84SYY119 pKa = 10.98KK120 pKa = 9.93RR121 pKa = 11.84RR122 pKa = 11.84STGGRR127 pKa = 11.84GGFAKK132 pKa = 10.17RR133 pKa = 11.84VLNVVTRR140 pKa = 11.84QNQAINTYY148 pKa = 8.0KK149 pKa = 10.2TSRR152 pKa = 11.84VDD154 pKa = 5.5FFDD157 pKa = 4.62LNQVDD162 pKa = 4.1GSTARR167 pKa = 11.84ALVCYY172 pKa = 9.8RR173 pKa = 11.84NCKK176 pKa = 10.22NYY178 pKa = 10.92ANGQAPGTALLMPTDD193 pKa = 4.84DD194 pKa = 3.93PHH196 pKa = 8.83ILTDD200 pKa = 3.33IANNIDD206 pKa = 3.21ASKK209 pKa = 6.81TTRR212 pKa = 11.84YY213 pKa = 7.52TRR215 pKa = 11.84LAYY218 pKa = 9.42KK219 pKa = 10.67VSGKK223 pKa = 7.22VTNMSNHH230 pKa = 6.15AADD233 pKa = 3.65VVHH236 pKa = 5.63YY237 pKa = 9.28RR238 pKa = 11.84CRR240 pKa = 11.84VRR242 pKa = 11.84KK243 pKa = 9.41DD244 pKa = 3.32VPPVQVGSTLGTFQTIVQSGFLGNNNGLGNAAVDD278 pKa = 4.27GVNQVQGASCTIPQAYY294 pKa = 10.2SLFEE298 pKa = 3.56NVQFVKK304 pKa = 10.33HH305 pKa = 5.84FKK307 pKa = 7.46ITKK310 pKa = 9.03VKK312 pKa = 7.12THH314 pKa = 5.82HH315 pKa = 6.49MEE317 pKa = 4.2PASSFKK323 pKa = 10.85LFYY326 pKa = 10.45KK327 pKa = 10.43LSKK330 pKa = 9.87PRR332 pKa = 11.84VVKK335 pKa = 10.99HH336 pKa = 6.26NDD338 pKa = 3.05YY339 pKa = 11.11TNLTNAYY346 pKa = 9.29LQEE349 pKa = 4.56LYY351 pKa = 10.55GGQYY355 pKa = 10.22FSVFVIRR362 pKa = 11.84GVLGYY367 pKa = 10.57DD368 pKa = 3.0KK369 pKa = 11.15SLAANLQLGYY379 pKa = 9.11TGADD383 pKa = 2.68IAYY386 pKa = 6.81EE387 pKa = 3.68QHH389 pKa = 7.13IEE391 pKa = 3.96VDD393 pKa = 4.03YY394 pKa = 9.63TWTVDD399 pKa = 3.1GSKK402 pKa = 10.83AFGMNQYY409 pKa = 7.51TTGLARR415 pKa = 11.84GHH417 pKa = 5.76VPAPIVEE424 pKa = 4.23NFNQNVVVYY433 pKa = 10.6NDD435 pKa = 3.68GQAKK439 pKa = 7.8FTTGSIGQTAWSTNPDD455 pKa = 4.48PIVEE459 pKa = 5.26DD460 pKa = 4.18GDD462 pKa = 3.67QQ463 pKa = 3.24
MM1 pKa = 6.94YY2 pKa = 10.17RR3 pKa = 11.84PRR5 pKa = 11.84RR6 pKa = 11.84TTNVGPTGSNPGLAIQRR23 pKa = 11.84LVFGSRR29 pKa = 11.84ARR31 pKa = 11.84EE32 pKa = 3.73EE33 pKa = 4.33DD34 pKa = 3.7SPAVSQGSQAKK45 pKa = 9.34KK46 pKa = 9.78RR47 pKa = 11.84RR48 pKa = 11.84LSRR51 pKa = 11.84SGGWAKK57 pKa = 10.53EE58 pKa = 3.6NRR60 pKa = 11.84TRR62 pKa = 11.84IPLYY66 pKa = 10.28SSDD69 pKa = 3.59MAKK72 pKa = 10.54SSIKK76 pKa = 10.3RR77 pKa = 11.84SGSRR81 pKa = 11.84IAKK84 pKa = 9.25RR85 pKa = 11.84RR86 pKa = 11.84YY87 pKa = 7.09TKK89 pKa = 10.45RR90 pKa = 11.84STKK93 pKa = 10.01GRR95 pKa = 11.84TRR97 pKa = 11.84RR98 pKa = 11.84PLSRR102 pKa = 11.84ARR104 pKa = 11.84PTRR107 pKa = 11.84KK108 pKa = 8.94FRR110 pKa = 11.84TRR112 pKa = 11.84KK113 pKa = 9.9SPARR117 pKa = 11.84SYY119 pKa = 10.98KK120 pKa = 9.93RR121 pKa = 11.84RR122 pKa = 11.84STGGRR127 pKa = 11.84GGFAKK132 pKa = 10.17RR133 pKa = 11.84VLNVVTRR140 pKa = 11.84QNQAINTYY148 pKa = 8.0KK149 pKa = 10.2TSRR152 pKa = 11.84VDD154 pKa = 5.5FFDD157 pKa = 4.62LNQVDD162 pKa = 4.1GSTARR167 pKa = 11.84ALVCYY172 pKa = 9.8RR173 pKa = 11.84NCKK176 pKa = 10.22NYY178 pKa = 10.92ANGQAPGTALLMPTDD193 pKa = 4.84DD194 pKa = 3.93PHH196 pKa = 8.83ILTDD200 pKa = 3.33IANNIDD206 pKa = 3.21ASKK209 pKa = 6.81TTRR212 pKa = 11.84YY213 pKa = 7.52TRR215 pKa = 11.84LAYY218 pKa = 9.42KK219 pKa = 10.67VSGKK223 pKa = 7.22VTNMSNHH230 pKa = 6.15AADD233 pKa = 3.65VVHH236 pKa = 5.63YY237 pKa = 9.28RR238 pKa = 11.84CRR240 pKa = 11.84VRR242 pKa = 11.84KK243 pKa = 9.41DD244 pKa = 3.32VPPVQVGSTLGTFQTIVQSGFLGNNNGLGNAAVDD278 pKa = 4.27GVNQVQGASCTIPQAYY294 pKa = 10.2SLFEE298 pKa = 3.56NVQFVKK304 pKa = 10.33HH305 pKa = 5.84FKK307 pKa = 7.46ITKK310 pKa = 9.03VKK312 pKa = 7.12THH314 pKa = 5.82HH315 pKa = 6.49MEE317 pKa = 4.2PASSFKK323 pKa = 10.85LFYY326 pKa = 10.45KK327 pKa = 10.43LSKK330 pKa = 9.87PRR332 pKa = 11.84VVKK335 pKa = 10.99HH336 pKa = 6.26NDD338 pKa = 3.05YY339 pKa = 11.11TNLTNAYY346 pKa = 9.29LQEE349 pKa = 4.56LYY351 pKa = 10.55GGQYY355 pKa = 10.22FSVFVIRR362 pKa = 11.84GVLGYY367 pKa = 10.57DD368 pKa = 3.0KK369 pKa = 11.15SLAANLQLGYY379 pKa = 9.11TGADD383 pKa = 2.68IAYY386 pKa = 6.81EE387 pKa = 3.68QHH389 pKa = 7.13IEE391 pKa = 3.96VDD393 pKa = 4.03YY394 pKa = 9.63TWTVDD399 pKa = 3.1GSKK402 pKa = 10.83AFGMNQYY409 pKa = 7.51TTGLARR415 pKa = 11.84GHH417 pKa = 5.76VPAPIVEE424 pKa = 4.23NFNQNVVVYY433 pKa = 10.6NDD435 pKa = 3.68GQAKK439 pKa = 7.8FTTGSIGQTAWSTNPDD455 pKa = 4.48PIVEE459 pKa = 5.26DD460 pKa = 4.18GDD462 pKa = 3.67QQ463 pKa = 3.24
Molecular weight: 51.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1127 |
316 |
463 |
375.7 |
41.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.808 ± 0.359 | 1.331 ± 0.665 |
5.768 ± 0.616 | 4.525 ± 1.165 |
2.484 ± 0.584 | 7.986 ± 0.651 |
2.928 ± 0.527 | 3.106 ± 0.288 |
5.146 ± 1.161 | 7.453 ± 1.427 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.42 ± 0.365 | 3.904 ± 1.286 |
4.348 ± 1.279 | 4.082 ± 0.506 |
8.075 ± 0.413 | 7.453 ± 0.873 |
6.921 ± 1.015 | 9.76 ± 2.667 |
1.331 ± 0.698 | 4.17 ± 1.072 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
