
Deer atadenovirus A
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Preplasmiviricota; Tectiliviricetes; Rowavirales; Adenoviridae; Atadenovirus
Average proteome isoelectric point is 6.85
Get precalculated fractions of proteins
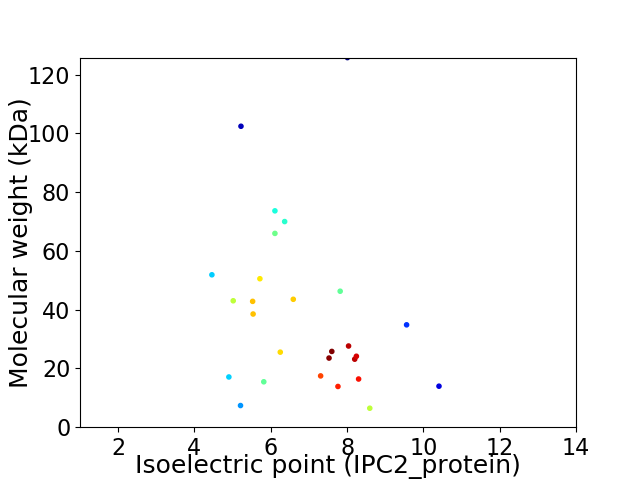
Virtual 2D-PAGE plot for 27 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A515MFS5|A0A515MFS5_9ADEN E4.2 OS=Deer atadenovirus A OX=2169706 PE=4 SV=1
MM1 pKa = 7.43KK2 pKa = 10.04RR3 pKa = 11.84ARR5 pKa = 11.84WDD7 pKa = 3.19PVYY10 pKa = 10.46PFSEE14 pKa = 4.7DD15 pKa = 3.13KK16 pKa = 10.99LIPLPPFIQAGTGLEE31 pKa = 4.17SEE33 pKa = 4.66GMILSLNFTDD43 pKa = 6.32PITINPTGSLTIKK56 pKa = 9.76IGPGIEE62 pKa = 3.95INEE65 pKa = 4.21NGQLTATAQALIAEE79 pKa = 4.64SPLKK83 pKa = 10.42KK84 pKa = 10.07EE85 pKa = 3.95NYY87 pKa = 8.67ILKK90 pKa = 9.66IQTNNTLVTEE100 pKa = 4.39DD101 pKa = 4.73DD102 pKa = 3.75KK103 pKa = 11.63MGIPDD108 pKa = 4.43PLEE111 pKa = 3.99PLNIEE116 pKa = 4.21SSGQLSLNFDD126 pKa = 3.27KK127 pKa = 11.2GFQLFEE133 pKa = 4.65GKK135 pKa = 10.53LSLYY139 pKa = 10.34LDD141 pKa = 3.4NSLFIKK147 pKa = 10.61NEE149 pKa = 3.8KK150 pKa = 10.37LSLKK154 pKa = 10.41EE155 pKa = 4.12PIEE158 pKa = 3.8PLIINSQGQLNLNYY172 pKa = 10.47DD173 pKa = 3.71NSLVLQDD180 pKa = 4.6HH181 pKa = 6.62KK182 pKa = 11.44LSVPEE187 pKa = 4.05PLEE190 pKa = 4.11PLDD193 pKa = 5.31LEE195 pKa = 4.66STGQLALSINKK206 pKa = 8.81GFQISDD212 pKa = 3.03GRR214 pKa = 11.84LTLDD218 pKa = 3.43IANCFQYY225 pKa = 11.13NSNKK229 pKa = 8.7LTLTNPTTPIVIKK242 pKa = 8.73PTGEE246 pKa = 3.29ISLNIGEE253 pKa = 4.82GLTVDD258 pKa = 4.54EE259 pKa = 6.02GNLTLNTNDD268 pKa = 3.51TLQIEE273 pKa = 4.79DD274 pKa = 3.92GQITLTDD281 pKa = 3.42PMEE284 pKa = 4.47PLNLEE289 pKa = 3.9MGQLVLNVDD298 pKa = 3.55NVFKK302 pKa = 11.37VEE304 pKa = 3.84NGKK307 pKa = 10.37LGLNVNSPMSINNNNLNINLGQGLEE332 pKa = 4.09IKK334 pKa = 10.47QGALTPKK341 pKa = 9.84ISSPLIFNSSGEE353 pKa = 4.26ITTSTTQGSQIIPFDD368 pKa = 3.77AYY370 pKa = 10.4EE371 pKa = 4.73LVLMWQKK378 pKa = 10.92FLTTRR383 pKa = 11.84HH384 pKa = 6.1AIYY387 pKa = 10.58VLHH390 pKa = 6.36CTSFIPDD397 pKa = 3.63TSVDD401 pKa = 4.02TITFTPNDD409 pKa = 3.71GLKK412 pKa = 10.76AFLSTDD418 pKa = 3.28SPLFTAAFQQVGNEE432 pKa = 3.92VKK434 pKa = 10.07TIGVKK439 pKa = 10.65FSKK442 pKa = 10.57ISDD445 pKa = 3.63TEE447 pKa = 4.06VQIKK451 pKa = 8.94YY452 pKa = 10.38SQMLTTQVILSSWTATTALFVV473 pKa = 3.62
MM1 pKa = 7.43KK2 pKa = 10.04RR3 pKa = 11.84ARR5 pKa = 11.84WDD7 pKa = 3.19PVYY10 pKa = 10.46PFSEE14 pKa = 4.7DD15 pKa = 3.13KK16 pKa = 10.99LIPLPPFIQAGTGLEE31 pKa = 4.17SEE33 pKa = 4.66GMILSLNFTDD43 pKa = 6.32PITINPTGSLTIKK56 pKa = 9.76IGPGIEE62 pKa = 3.95INEE65 pKa = 4.21NGQLTATAQALIAEE79 pKa = 4.64SPLKK83 pKa = 10.42KK84 pKa = 10.07EE85 pKa = 3.95NYY87 pKa = 8.67ILKK90 pKa = 9.66IQTNNTLVTEE100 pKa = 4.39DD101 pKa = 4.73DD102 pKa = 3.75KK103 pKa = 11.63MGIPDD108 pKa = 4.43PLEE111 pKa = 3.99PLNIEE116 pKa = 4.21SSGQLSLNFDD126 pKa = 3.27KK127 pKa = 11.2GFQLFEE133 pKa = 4.65GKK135 pKa = 10.53LSLYY139 pKa = 10.34LDD141 pKa = 3.4NSLFIKK147 pKa = 10.61NEE149 pKa = 3.8KK150 pKa = 10.37LSLKK154 pKa = 10.41EE155 pKa = 4.12PIEE158 pKa = 3.8PLIINSQGQLNLNYY172 pKa = 10.47DD173 pKa = 3.71NSLVLQDD180 pKa = 4.6HH181 pKa = 6.62KK182 pKa = 11.44LSVPEE187 pKa = 4.05PLEE190 pKa = 4.11PLDD193 pKa = 5.31LEE195 pKa = 4.66STGQLALSINKK206 pKa = 8.81GFQISDD212 pKa = 3.03GRR214 pKa = 11.84LTLDD218 pKa = 3.43IANCFQYY225 pKa = 11.13NSNKK229 pKa = 8.7LTLTNPTTPIVIKK242 pKa = 8.73PTGEE246 pKa = 3.29ISLNIGEE253 pKa = 4.82GLTVDD258 pKa = 4.54EE259 pKa = 6.02GNLTLNTNDD268 pKa = 3.51TLQIEE273 pKa = 4.79DD274 pKa = 3.92GQITLTDD281 pKa = 3.42PMEE284 pKa = 4.47PLNLEE289 pKa = 3.9MGQLVLNVDD298 pKa = 3.55NVFKK302 pKa = 11.37VEE304 pKa = 3.84NGKK307 pKa = 10.37LGLNVNSPMSINNNNLNINLGQGLEE332 pKa = 4.09IKK334 pKa = 10.47QGALTPKK341 pKa = 9.84ISSPLIFNSSGEE353 pKa = 4.26ITTSTTQGSQIIPFDD368 pKa = 3.77AYY370 pKa = 10.4EE371 pKa = 4.73LVLMWQKK378 pKa = 10.92FLTTRR383 pKa = 11.84HH384 pKa = 6.1AIYY387 pKa = 10.58VLHH390 pKa = 6.36CTSFIPDD397 pKa = 3.63TSVDD401 pKa = 4.02TITFTPNDD409 pKa = 3.71GLKK412 pKa = 10.76AFLSTDD418 pKa = 3.28SPLFTAAFQQVGNEE432 pKa = 3.92VKK434 pKa = 10.07TIGVKK439 pKa = 10.65FSKK442 pKa = 10.57ISDD445 pKa = 3.63TEE447 pKa = 4.06VQIKK451 pKa = 8.94YY452 pKa = 10.38SQMLTTQVILSSWTATTALFVV473 pKa = 3.62
Molecular weight: 51.89 kDa
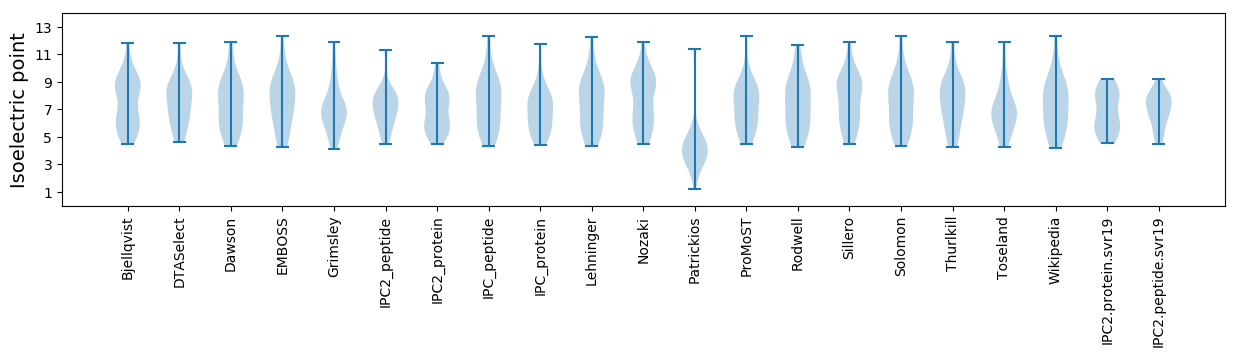
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A515MG01|A0A515MG01_9ADEN PVI OS=Deer atadenovirus A OX=2169706 PE=4 SV=1
MM1 pKa = 7.6EE2 pKa = 5.57SMPHH6 pKa = 7.36LYY8 pKa = 9.93TLMGGATRR16 pKa = 11.84KK17 pKa = 8.93KK18 pKa = 9.79RR19 pKa = 11.84KK20 pKa = 9.4RR21 pKa = 11.84RR22 pKa = 11.84TITRR26 pKa = 11.84KK27 pKa = 9.69RR28 pKa = 11.84NPIKK32 pKa = 10.63SSNRR36 pKa = 11.84KK37 pKa = 5.99ITRR40 pKa = 11.84GTRR43 pKa = 11.84RR44 pKa = 11.84LKK46 pKa = 10.29HH47 pKa = 4.58VRR49 pKa = 11.84IPIVSQYY56 pKa = 11.54LNIHH60 pKa = 6.85PPQALNKK67 pKa = 8.33QHH69 pKa = 6.37QPLFYY74 pKa = 10.59HH75 pKa = 7.05HH76 pKa = 7.16FNQHH80 pKa = 5.29LHH82 pKa = 5.89HH83 pKa = 6.59QPNQPLVGTSYY94 pKa = 11.0PMPSTSIFIHH104 pKa = 6.88PPTINNSFTKK114 pKa = 10.41NVSLKK119 pKa = 10.07RR120 pKa = 11.84DD121 pKa = 3.66HH122 pKa = 6.94QPDD125 pKa = 4.46EE126 pKa = 4.01IQVFHH131 pKa = 7.61DD132 pKa = 4.02AQDD135 pKa = 3.14HH136 pKa = 5.61FEE138 pKa = 4.14IKK140 pKa = 9.77RR141 pKa = 11.84TDD143 pKa = 3.38GEE145 pKa = 4.07NDD147 pKa = 2.92SDD149 pKa = 4.37INAKK153 pKa = 10.15KK154 pKa = 9.98KK155 pKa = 9.58WSLEE159 pKa = 3.96DD160 pKa = 3.38VLSFLQKK167 pKa = 10.3VPHH170 pKa = 5.56QLRR173 pKa = 11.84KK174 pKa = 9.87IILTSLFGATIGLLFDD190 pKa = 5.2LLLGGPWGLTTRR202 pKa = 11.84LLRR205 pKa = 11.84LIISLVPGGNIFLTALDD222 pKa = 3.8GLGYY226 pKa = 11.03LLGKK230 pKa = 9.3SANPFNIAYY239 pKa = 10.14DD240 pKa = 3.59PDD242 pKa = 3.72FQNFGNIIQSKK253 pKa = 8.87MNDD256 pKa = 3.11RR257 pKa = 11.84LAEE260 pKa = 4.3DD261 pKa = 3.53VAKK264 pKa = 10.51AAEE267 pKa = 4.03EE268 pKa = 4.0QIGNGFMRR276 pKa = 11.84TLASLLSAAASAGTHH291 pKa = 6.24LKK293 pKa = 10.32LALPAIPIAAIRR305 pKa = 11.84PFQRR309 pKa = 4.05
MM1 pKa = 7.6EE2 pKa = 5.57SMPHH6 pKa = 7.36LYY8 pKa = 9.93TLMGGATRR16 pKa = 11.84KK17 pKa = 8.93KK18 pKa = 9.79RR19 pKa = 11.84KK20 pKa = 9.4RR21 pKa = 11.84RR22 pKa = 11.84TITRR26 pKa = 11.84KK27 pKa = 9.69RR28 pKa = 11.84NPIKK32 pKa = 10.63SSNRR36 pKa = 11.84KK37 pKa = 5.99ITRR40 pKa = 11.84GTRR43 pKa = 11.84RR44 pKa = 11.84LKK46 pKa = 10.29HH47 pKa = 4.58VRR49 pKa = 11.84IPIVSQYY56 pKa = 11.54LNIHH60 pKa = 6.85PPQALNKK67 pKa = 8.33QHH69 pKa = 6.37QPLFYY74 pKa = 10.59HH75 pKa = 7.05HH76 pKa = 7.16FNQHH80 pKa = 5.29LHH82 pKa = 5.89HH83 pKa = 6.59QPNQPLVGTSYY94 pKa = 11.0PMPSTSIFIHH104 pKa = 6.88PPTINNSFTKK114 pKa = 10.41NVSLKK119 pKa = 10.07RR120 pKa = 11.84DD121 pKa = 3.66HH122 pKa = 6.94QPDD125 pKa = 4.46EE126 pKa = 4.01IQVFHH131 pKa = 7.61DD132 pKa = 4.02AQDD135 pKa = 3.14HH136 pKa = 5.61FEE138 pKa = 4.14IKK140 pKa = 9.77RR141 pKa = 11.84TDD143 pKa = 3.38GEE145 pKa = 4.07NDD147 pKa = 2.92SDD149 pKa = 4.37INAKK153 pKa = 10.15KK154 pKa = 9.98KK155 pKa = 9.58WSLEE159 pKa = 3.96DD160 pKa = 3.38VLSFLQKK167 pKa = 10.3VPHH170 pKa = 5.56QLRR173 pKa = 11.84KK174 pKa = 9.87IILTSLFGATIGLLFDD190 pKa = 5.2LLLGGPWGLTTRR202 pKa = 11.84LLRR205 pKa = 11.84LIISLVPGGNIFLTALDD222 pKa = 3.8GLGYY226 pKa = 11.03LLGKK230 pKa = 9.3SANPFNIAYY239 pKa = 10.14DD240 pKa = 3.59PDD242 pKa = 3.72FQNFGNIIQSKK253 pKa = 8.87MNDD256 pKa = 3.11RR257 pKa = 11.84LAEE260 pKa = 4.3DD261 pKa = 3.53VAKK264 pKa = 10.51AAEE267 pKa = 4.03EE268 pKa = 4.0QIGNGFMRR276 pKa = 11.84TLASLLSAAASAGTHH291 pKa = 6.24LKK293 pKa = 10.32LALPAIPIAAIRR305 pKa = 11.84PFQRR309 pKa = 4.05
Molecular weight: 34.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
9146 |
54 |
1075 |
338.7 |
38.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.855 ± 0.309 | 2.252 ± 0.462 |
4.855 ± 0.227 | 5.839 ± 0.364 |
5.576 ± 0.27 | 5.084 ± 0.374 |
2.405 ± 0.284 | 6.845 ± 0.393 |
6.615 ± 0.63 | 9.48 ± 0.499 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.045 ± 0.156 | 6.571 ± 0.401 |
5.019 ± 0.283 | 4.516 ± 0.245 |
4.428 ± 0.476 | 7.194 ± 0.345 |
6.254 ± 0.371 | 4.778 ± 0.318 |
1.498 ± 0.152 | 3.892 ± 0.319 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
