
Roseomonas cervicalis ATCC 49957
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Acetobacteraceae; Roseomonas; Roseomonas cervicalis
Average proteome isoelectric point is 7.11
Get precalculated fractions of proteins
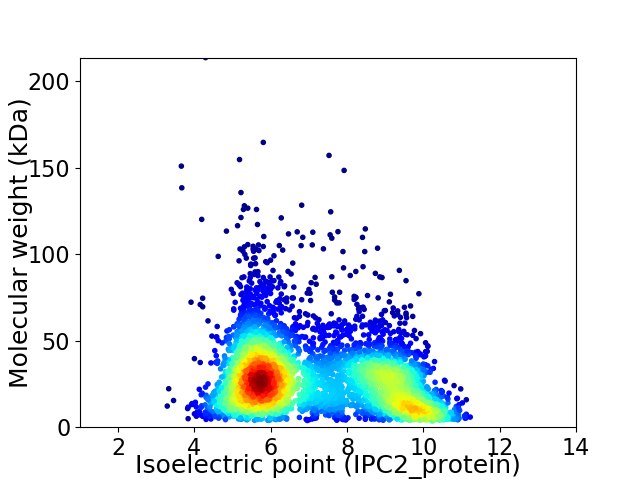
Virtual 2D-PAGE plot for 4774 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
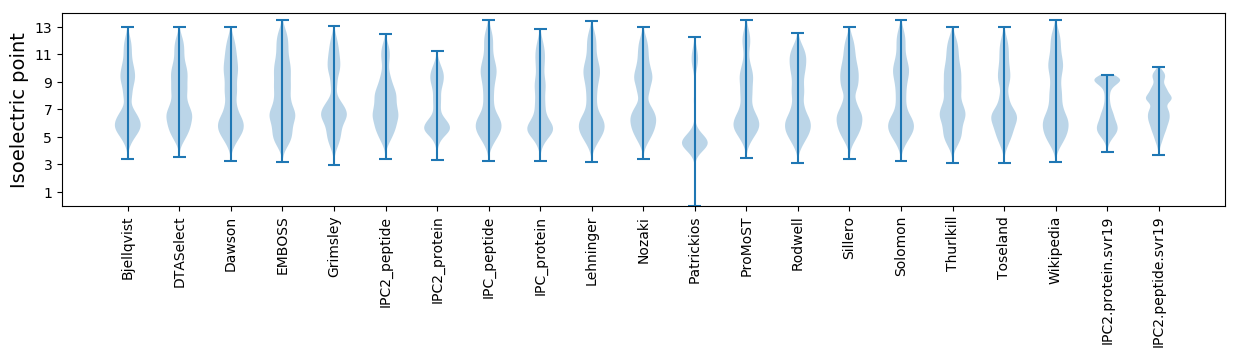
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D5RIW8|D5RIW8_9PROT Carbonic anhydrase OS=Roseomonas cervicalis ATCC 49957 OX=525371 GN=cah PE=3 SV=1
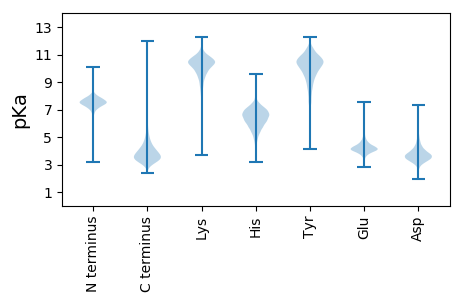
MM1 pKa = 7.5AVLIGTPGNDD11 pKa = 2.81RR12 pKa = 11.84LIAPDD17 pKa = 3.41ARR19 pKa = 11.84EE20 pKa = 3.99NDD22 pKa = 4.41SIAGDD27 pKa = 3.71AGDD30 pKa = 4.86DD31 pKa = 4.06FIEE34 pKa = 4.84ARR36 pKa = 11.84GGDD39 pKa = 3.9DD40 pKa = 5.18RR41 pKa = 11.84ITPGPGNDD49 pKa = 2.98RR50 pKa = 11.84VEE52 pKa = 4.71GGDD55 pKa = 3.52GRR57 pKa = 11.84DD58 pKa = 3.44TVIVSGDD65 pKa = 3.0ISQTEE70 pKa = 4.43VYY72 pKa = 9.82RR73 pKa = 11.84YY74 pKa = 10.25NNEE77 pKa = 3.76GVLRR81 pKa = 11.84GPDD84 pKa = 3.66GVDD87 pKa = 2.79TLLDD91 pKa = 3.53VEE93 pKa = 5.15AVQFTGVGGTLEE105 pKa = 4.01MSDD108 pKa = 3.72ANSFLSYY115 pKa = 10.79SYY117 pKa = 10.14IASYY121 pKa = 11.53GDD123 pKa = 3.26LTEE126 pKa = 5.76AYY128 pKa = 9.78GADD131 pKa = 3.52AGAGWRR137 pKa = 11.84HH138 pKa = 5.18FRR140 pKa = 11.84DD141 pKa = 4.68FGAVEE146 pKa = 3.9GRR148 pKa = 11.84EE149 pKa = 3.67ITFNGNAYY157 pKa = 10.04LAANTDD163 pKa = 3.61VLSALGANADD173 pKa = 3.67EE174 pKa = 4.8SGARR178 pKa = 11.84HH179 pKa = 4.89YY180 pKa = 11.18LEE182 pKa = 4.01YY183 pKa = 11.18GRR185 pKa = 11.84FEE187 pKa = 4.03GRR189 pKa = 11.84TTEE192 pKa = 4.45FAALSYY198 pKa = 8.42TASYY202 pKa = 10.93GEE204 pKa = 5.1LIDD207 pKa = 5.37SFGTDD212 pKa = 3.86TIAATAHH219 pKa = 6.08FVQEE223 pKa = 4.32GFNEE227 pKa = 3.93GRR229 pKa = 11.84GISFNGLEE237 pKa = 4.3YY238 pKa = 10.34VASYY242 pKa = 11.23GDD244 pKa = 4.91LIDD247 pKa = 5.11AYY249 pKa = 10.55GDD251 pKa = 3.4AEE253 pKa = 4.3RR254 pKa = 11.84PFDD257 pKa = 4.99LGEE260 pKa = 5.04DD261 pKa = 4.04GAGHH265 pKa = 6.82YY266 pKa = 9.18IQYY269 pKa = 10.6GRR271 pKa = 11.84GEE273 pKa = 4.15GRR275 pKa = 11.84EE276 pKa = 4.04TTFDD280 pKa = 3.22GLQYY284 pKa = 8.82MASYY288 pKa = 10.8GDD290 pKa = 3.65VIEE293 pKa = 4.77AFRR296 pKa = 11.84DD297 pKa = 3.67STDD300 pKa = 2.66AGAYY304 pKa = 7.13DD305 pKa = 4.25TIGALHH311 pKa = 6.9YY312 pKa = 10.59IRR314 pKa = 11.84DD315 pKa = 3.82GFGEE319 pKa = 4.07EE320 pKa = 3.57RR321 pKa = 11.84VADD324 pKa = 4.17RR325 pKa = 11.84FNEE328 pKa = 3.92QSYY331 pKa = 10.52AAANGDD337 pKa = 3.73LAEE340 pKa = 4.8AGITSADD347 pKa = 3.28ALALHH352 pKa = 7.57WIQYY356 pKa = 9.77GYY358 pKa = 11.13EE359 pKa = 3.9KK360 pKa = 10.73GRR362 pKa = 11.84AGAYY366 pKa = 10.32DD367 pKa = 3.67PVIAA371 pKa = 5.43
MM1 pKa = 7.5AVLIGTPGNDD11 pKa = 2.81RR12 pKa = 11.84LIAPDD17 pKa = 3.41ARR19 pKa = 11.84EE20 pKa = 3.99NDD22 pKa = 4.41SIAGDD27 pKa = 3.71AGDD30 pKa = 4.86DD31 pKa = 4.06FIEE34 pKa = 4.84ARR36 pKa = 11.84GGDD39 pKa = 3.9DD40 pKa = 5.18RR41 pKa = 11.84ITPGPGNDD49 pKa = 2.98RR50 pKa = 11.84VEE52 pKa = 4.71GGDD55 pKa = 3.52GRR57 pKa = 11.84DD58 pKa = 3.44TVIVSGDD65 pKa = 3.0ISQTEE70 pKa = 4.43VYY72 pKa = 9.82RR73 pKa = 11.84YY74 pKa = 10.25NNEE77 pKa = 3.76GVLRR81 pKa = 11.84GPDD84 pKa = 3.66GVDD87 pKa = 2.79TLLDD91 pKa = 3.53VEE93 pKa = 5.15AVQFTGVGGTLEE105 pKa = 4.01MSDD108 pKa = 3.72ANSFLSYY115 pKa = 10.79SYY117 pKa = 10.14IASYY121 pKa = 11.53GDD123 pKa = 3.26LTEE126 pKa = 5.76AYY128 pKa = 9.78GADD131 pKa = 3.52AGAGWRR137 pKa = 11.84HH138 pKa = 5.18FRR140 pKa = 11.84DD141 pKa = 4.68FGAVEE146 pKa = 3.9GRR148 pKa = 11.84EE149 pKa = 3.67ITFNGNAYY157 pKa = 10.04LAANTDD163 pKa = 3.61VLSALGANADD173 pKa = 3.67EE174 pKa = 4.8SGARR178 pKa = 11.84HH179 pKa = 4.89YY180 pKa = 11.18LEE182 pKa = 4.01YY183 pKa = 11.18GRR185 pKa = 11.84FEE187 pKa = 4.03GRR189 pKa = 11.84TTEE192 pKa = 4.45FAALSYY198 pKa = 8.42TASYY202 pKa = 10.93GEE204 pKa = 5.1LIDD207 pKa = 5.37SFGTDD212 pKa = 3.86TIAATAHH219 pKa = 6.08FVQEE223 pKa = 4.32GFNEE227 pKa = 3.93GRR229 pKa = 11.84GISFNGLEE237 pKa = 4.3YY238 pKa = 10.34VASYY242 pKa = 11.23GDD244 pKa = 4.91LIDD247 pKa = 5.11AYY249 pKa = 10.55GDD251 pKa = 3.4AEE253 pKa = 4.3RR254 pKa = 11.84PFDD257 pKa = 4.99LGEE260 pKa = 5.04DD261 pKa = 4.04GAGHH265 pKa = 6.82YY266 pKa = 9.18IQYY269 pKa = 10.6GRR271 pKa = 11.84GEE273 pKa = 4.15GRR275 pKa = 11.84EE276 pKa = 4.04TTFDD280 pKa = 3.22GLQYY284 pKa = 8.82MASYY288 pKa = 10.8GDD290 pKa = 3.65VIEE293 pKa = 4.77AFRR296 pKa = 11.84DD297 pKa = 3.67STDD300 pKa = 2.66AGAYY304 pKa = 7.13DD305 pKa = 4.25TIGALHH311 pKa = 6.9YY312 pKa = 10.59IRR314 pKa = 11.84DD315 pKa = 3.82GFGEE319 pKa = 4.07EE320 pKa = 3.57RR321 pKa = 11.84VADD324 pKa = 4.17RR325 pKa = 11.84FNEE328 pKa = 3.92QSYY331 pKa = 10.52AAANGDD337 pKa = 3.73LAEE340 pKa = 4.8AGITSADD347 pKa = 3.28ALALHH352 pKa = 7.57WIQYY356 pKa = 9.77GYY358 pKa = 11.13EE359 pKa = 3.9KK360 pKa = 10.73GRR362 pKa = 11.84AGAYY366 pKa = 10.32DD367 pKa = 3.67PVIAA371 pKa = 5.43
Molecular weight: 39.62 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D5RJG8|D5RJG8_9PROT Uncharacterized protein (Fragment) OS=Roseomonas cervicalis ATCC 49957 OX=525371 GN=HMPREF0731_1228 PE=4 SV=1
MM1 pKa = 7.36GRR3 pKa = 11.84GGTLGRR9 pKa = 11.84AAAVPWRR16 pKa = 11.84SGARR20 pKa = 11.84LRR22 pKa = 11.84AAAIRR27 pKa = 11.84WGGKK31 pKa = 9.41AGSGAIPWRR40 pKa = 11.84GRR42 pKa = 11.84RR43 pKa = 11.84PRR45 pKa = 11.84AGRR48 pKa = 11.84QAWHH52 pKa = 6.99PAASS56 pKa = 3.41
MM1 pKa = 7.36GRR3 pKa = 11.84GGTLGRR9 pKa = 11.84AAAVPWRR16 pKa = 11.84SGARR20 pKa = 11.84LRR22 pKa = 11.84AAAIRR27 pKa = 11.84WGGKK31 pKa = 9.41AGSGAIPWRR40 pKa = 11.84GRR42 pKa = 11.84RR43 pKa = 11.84PRR45 pKa = 11.84AGRR48 pKa = 11.84QAWHH52 pKa = 6.99PAASS56 pKa = 3.41
Molecular weight: 5.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1327154 |
39 |
2039 |
278.0 |
29.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
15.034 ± 0.058 | 0.815 ± 0.009 |
4.747 ± 0.028 | 5.728 ± 0.038 |
3.198 ± 0.021 | 9.444 ± 0.038 |
1.981 ± 0.018 | 3.933 ± 0.026 |
1.952 ± 0.029 | 11.628 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.307 ± 0.017 | 1.929 ± 0.019 |
6.302 ± 0.029 | 3.339 ± 0.025 |
8.53 ± 0.042 | 4.467 ± 0.024 |
4.602 ± 0.025 | 6.859 ± 0.029 |
1.447 ± 0.016 | 1.758 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
