
Tessaracoccus flavus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Propionibacteriaceae; Tessaracoccus
Average proteome isoelectric point is 5.99
Get precalculated fractions of proteins
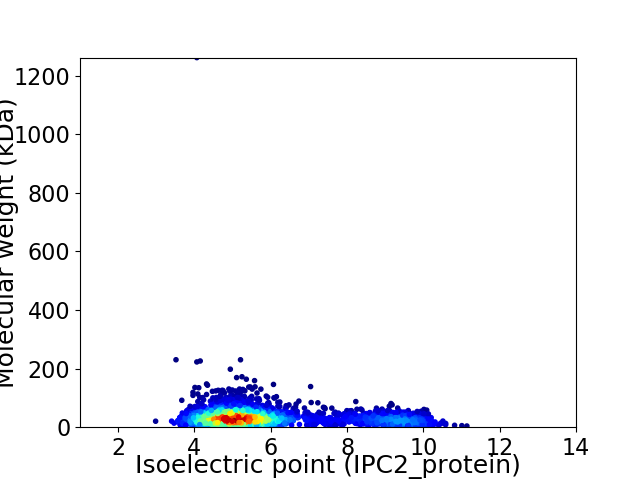
Virtual 2D-PAGE plot for 2784 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
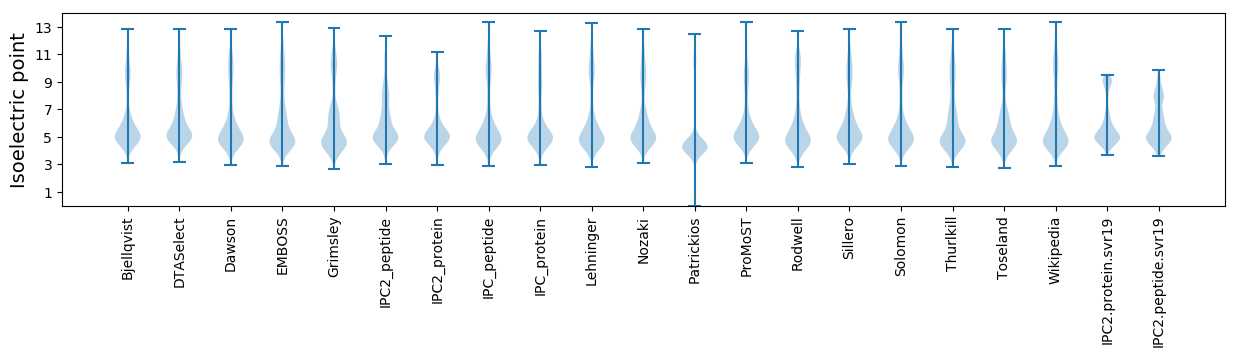
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Q2CGY6|A0A1Q2CGY6_9ACTN Predicted ATP-dependent carboligase ATP-grasp superfamily OS=Tessaracoccus flavus OX=1610493 GN=RPIT_11610 PE=4 SV=1
MM1 pKa = 7.22ITFKK5 pKa = 10.97KK6 pKa = 10.59ALTATALASALLLTACGGDD25 pKa = 3.76GDD27 pKa = 5.22DD28 pKa = 4.67PAPGATDD35 pKa = 3.41PAMTTPAATSGEE47 pKa = 4.47TTGGEE52 pKa = 4.03TTGAGEE58 pKa = 4.29APAASGTINFFTDD71 pKa = 2.93KK72 pKa = 10.92AAWEE76 pKa = 4.11PSFEE80 pKa = 4.21EE81 pKa = 4.11MNAASGDD88 pKa = 3.27IDD90 pKa = 5.44LEE92 pKa = 4.18FTGYY96 pKa = 10.11SDD98 pKa = 3.4PVAYY102 pKa = 8.23DD103 pKa = 3.02TYY105 pKa = 10.73IRR107 pKa = 11.84QAFRR111 pKa = 11.84TDD113 pKa = 3.51DD114 pKa = 3.86KK115 pKa = 11.38PDD117 pKa = 3.27LFTWHH122 pKa = 6.9TGGKK126 pKa = 8.59LQEE129 pKa = 4.31LAEE132 pKa = 4.15QGLVAEE138 pKa = 4.98TTDD141 pKa = 2.62IWAEE145 pKa = 3.97AEE147 pKa = 4.31SEE149 pKa = 4.31GLVPEE154 pKa = 4.41GLKK157 pKa = 10.55DD158 pKa = 3.18QYY160 pKa = 11.38TYY162 pKa = 11.08DD163 pKa = 3.43GKK165 pKa = 10.78QYY167 pKa = 10.64CVPLNVAYY175 pKa = 9.29WAVYY179 pKa = 9.01YY180 pKa = 10.53NKK182 pKa = 10.22QIFDD186 pKa = 4.64DD187 pKa = 4.42NGLTPPTTWEE197 pKa = 3.77EE198 pKa = 4.43MEE200 pKa = 4.55TVMQTLRR207 pKa = 11.84DD208 pKa = 3.54AGVTPFHH215 pKa = 6.69QMNIIFEE222 pKa = 4.2FVYY225 pKa = 9.92FQALLAGQDD234 pKa = 3.34PDD236 pKa = 3.82TYY238 pKa = 11.26LGILDD243 pKa = 4.58GSASYY248 pKa = 10.69TDD250 pKa = 3.48EE251 pKa = 4.48DD252 pKa = 4.51VNVAVDD258 pKa = 3.11KK259 pKa = 10.26WEE261 pKa = 3.83EE262 pKa = 3.91MINRR266 pKa = 11.84GDD268 pKa = 4.03FIDD271 pKa = 4.47PGVTGDD277 pKa = 3.86PQALLMNGDD286 pKa = 2.8VAMAYY291 pKa = 10.11FGTFFTGQLTAIDD304 pKa = 3.95AVSGEE309 pKa = 4.41DD310 pKa = 3.18YY311 pKa = 11.14GIFMFPNLNPDD322 pKa = 3.46TEE324 pKa = 4.4EE325 pKa = 3.98QPMILEE331 pKa = 4.51TGPLCVGAGSEE342 pKa = 4.5NEE344 pKa = 4.16DD345 pKa = 3.33TALAYY350 pKa = 10.26SKK352 pKa = 9.62WWLTTDD358 pKa = 3.9AQTAWSQSRR367 pKa = 11.84GDD369 pKa = 3.36VSFNPNVEE377 pKa = 3.96IEE379 pKa = 4.21DD380 pKa = 4.39PEE382 pKa = 4.69LSALVEE388 pKa = 4.07AVNDD392 pKa = 3.88GTRR395 pKa = 11.84ILPRR399 pKa = 11.84YY400 pKa = 9.88LEE402 pKa = 4.73GSPQPVYY409 pKa = 10.6QLSTEE414 pKa = 4.05LFGAFVTSPSDD425 pKa = 3.26PRR427 pKa = 11.84GMQEE431 pKa = 4.13RR432 pKa = 11.84LQAAADD438 pKa = 4.48DD439 pKa = 4.27YY440 pKa = 10.85WAQQQ444 pKa = 3.0
MM1 pKa = 7.22ITFKK5 pKa = 10.97KK6 pKa = 10.59ALTATALASALLLTACGGDD25 pKa = 3.76GDD27 pKa = 5.22DD28 pKa = 4.67PAPGATDD35 pKa = 3.41PAMTTPAATSGEE47 pKa = 4.47TTGGEE52 pKa = 4.03TTGAGEE58 pKa = 4.29APAASGTINFFTDD71 pKa = 2.93KK72 pKa = 10.92AAWEE76 pKa = 4.11PSFEE80 pKa = 4.21EE81 pKa = 4.11MNAASGDD88 pKa = 3.27IDD90 pKa = 5.44LEE92 pKa = 4.18FTGYY96 pKa = 10.11SDD98 pKa = 3.4PVAYY102 pKa = 8.23DD103 pKa = 3.02TYY105 pKa = 10.73IRR107 pKa = 11.84QAFRR111 pKa = 11.84TDD113 pKa = 3.51DD114 pKa = 3.86KK115 pKa = 11.38PDD117 pKa = 3.27LFTWHH122 pKa = 6.9TGGKK126 pKa = 8.59LQEE129 pKa = 4.31LAEE132 pKa = 4.15QGLVAEE138 pKa = 4.98TTDD141 pKa = 2.62IWAEE145 pKa = 3.97AEE147 pKa = 4.31SEE149 pKa = 4.31GLVPEE154 pKa = 4.41GLKK157 pKa = 10.55DD158 pKa = 3.18QYY160 pKa = 11.38TYY162 pKa = 11.08DD163 pKa = 3.43GKK165 pKa = 10.78QYY167 pKa = 10.64CVPLNVAYY175 pKa = 9.29WAVYY179 pKa = 9.01YY180 pKa = 10.53NKK182 pKa = 10.22QIFDD186 pKa = 4.64DD187 pKa = 4.42NGLTPPTTWEE197 pKa = 3.77EE198 pKa = 4.43MEE200 pKa = 4.55TVMQTLRR207 pKa = 11.84DD208 pKa = 3.54AGVTPFHH215 pKa = 6.69QMNIIFEE222 pKa = 4.2FVYY225 pKa = 9.92FQALLAGQDD234 pKa = 3.34PDD236 pKa = 3.82TYY238 pKa = 11.26LGILDD243 pKa = 4.58GSASYY248 pKa = 10.69TDD250 pKa = 3.48EE251 pKa = 4.48DD252 pKa = 4.51VNVAVDD258 pKa = 3.11KK259 pKa = 10.26WEE261 pKa = 3.83EE262 pKa = 3.91MINRR266 pKa = 11.84GDD268 pKa = 4.03FIDD271 pKa = 4.47PGVTGDD277 pKa = 3.86PQALLMNGDD286 pKa = 2.8VAMAYY291 pKa = 10.11FGTFFTGQLTAIDD304 pKa = 3.95AVSGEE309 pKa = 4.41DD310 pKa = 3.18YY311 pKa = 11.14GIFMFPNLNPDD322 pKa = 3.46TEE324 pKa = 4.4EE325 pKa = 3.98QPMILEE331 pKa = 4.51TGPLCVGAGSEE342 pKa = 4.5NEE344 pKa = 4.16DD345 pKa = 3.33TALAYY350 pKa = 10.26SKK352 pKa = 9.62WWLTTDD358 pKa = 3.9AQTAWSQSRR367 pKa = 11.84GDD369 pKa = 3.36VSFNPNVEE377 pKa = 3.96IEE379 pKa = 4.21DD380 pKa = 4.39PEE382 pKa = 4.69LSALVEE388 pKa = 4.07AVNDD392 pKa = 3.88GTRR395 pKa = 11.84ILPRR399 pKa = 11.84YY400 pKa = 9.88LEE402 pKa = 4.73GSPQPVYY409 pKa = 10.6QLSTEE414 pKa = 4.05LFGAFVTSPSDD425 pKa = 3.26PRR427 pKa = 11.84GMQEE431 pKa = 4.13RR432 pKa = 11.84LQAAADD438 pKa = 4.48DD439 pKa = 4.27YY440 pKa = 10.85WAQQQ444 pKa = 3.0
Molecular weight: 48.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Q2CC96|A0A1Q2CC96_9ACTN Uncharacterized protein OS=Tessaracoccus flavus OX=1610493 GN=RPIT_02000 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.55LLKK22 pKa = 8.44KK23 pKa = 9.1TRR25 pKa = 11.84IQRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84AGKK33 pKa = 9.79
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.55LLKK22 pKa = 8.44KK23 pKa = 9.1TRR25 pKa = 11.84IQRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84AGKK33 pKa = 9.79
Molecular weight: 4.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
923675 |
33 |
12360 |
331.8 |
35.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.391 ± 0.065 | 0.675 ± 0.016 |
6.359 ± 0.041 | 5.908 ± 0.05 |
3.082 ± 0.031 | 8.849 ± 0.065 |
2.116 ± 0.03 | 4.291 ± 0.037 |
2.223 ± 0.032 | 10.227 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.043 ± 0.027 | 2.149 ± 0.027 |
5.421 ± 0.038 | 2.979 ± 0.023 |
7.141 ± 0.074 | 5.609 ± 0.034 |
6.079 ± 0.076 | 8.818 ± 0.043 |
1.568 ± 0.022 | 2.073 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
