
Alteromonadaceae bacterium M269
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Alteromonadaceae; unclassified Alteromonadaceae
Average proteome isoelectric point is 5.96
Get precalculated fractions of proteins
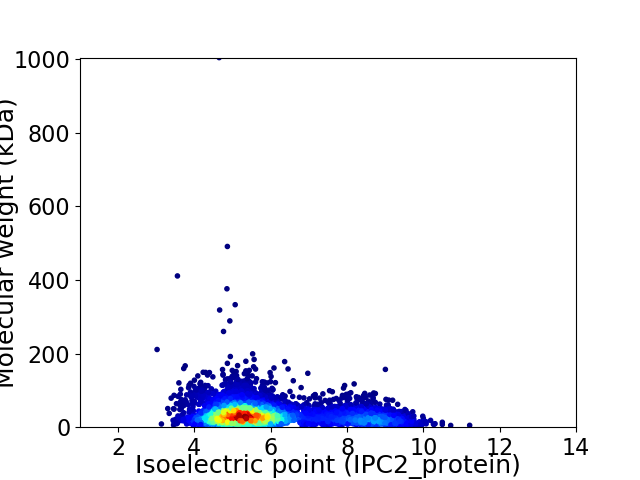
Virtual 2D-PAGE plot for 4871 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V2ZHA3|A0A4V2ZHA3_9ALTE Uncharacterized protein OS=Alteromonadaceae bacterium M269 OX=2546219 GN=EYS14_05510 PE=4 SV=1
MM1 pKa = 6.48TTLSIGLIMAFQSQADD17 pKa = 3.74QTQMVDD23 pKa = 3.08QPGSVLVGYY32 pKa = 7.51WHH34 pKa = 6.7NFCNADD40 pKa = 3.35GYY42 pKa = 11.21QRR44 pKa = 11.84GNAPCISLTSTNPLYY59 pKa = 11.1NVVNVSFMRR68 pKa = 11.84VNDD71 pKa = 3.82TSQGRR76 pKa = 11.84IPTFEE81 pKa = 5.09LDD83 pKa = 3.26PGAGLSEE90 pKa = 4.06QEE92 pKa = 4.93FINQIGQLNAQGRR105 pKa = 11.84SVLIALGGADD115 pKa = 3.95AHH117 pKa = 7.34IEE119 pKa = 4.04LQTGDD124 pKa = 3.59EE125 pKa = 4.0QAFADD130 pKa = 4.53EE131 pKa = 4.98IIRR134 pKa = 11.84LVEE137 pKa = 4.05TYY139 pKa = 10.83GFDD142 pKa = 4.08GLDD145 pKa = 3.15IDD147 pKa = 5.55LEE149 pKa = 4.31QAAITARR156 pKa = 11.84DD157 pKa = 3.8NQTVIPAALRR167 pKa = 11.84MVKK170 pKa = 10.09DD171 pKa = 3.78HH172 pKa = 6.74YY173 pKa = 10.58RR174 pKa = 11.84AQGKK178 pKa = 9.06NFMITMAPEE187 pKa = 4.17FPYY190 pKa = 9.93LTGAGAYY197 pKa = 9.57VPYY200 pKa = 10.58LQGLEE205 pKa = 4.24GYY207 pKa = 10.35YY208 pKa = 10.88DD209 pKa = 4.57FINPQFYY216 pKa = 10.67NQGGDD221 pKa = 3.18GLYY224 pKa = 9.73IDD226 pKa = 4.17GVGWIAQNNDD236 pKa = 2.95ALKK239 pKa = 10.84EE240 pKa = 3.9EE241 pKa = 4.81FIYY244 pKa = 10.84YY245 pKa = 10.05ISDD248 pKa = 4.59AIANGSRR255 pKa = 11.84GFAQIPAEE263 pKa = 3.8RR264 pKa = 11.84LVFGIPANNDD274 pKa = 2.72AAATGFVADD283 pKa = 4.15PADD286 pKa = 4.28LYY288 pKa = 11.09RR289 pKa = 11.84AFGRR293 pKa = 11.84MRR295 pKa = 11.84EE296 pKa = 4.01QGQPLRR302 pKa = 11.84GIMTWSINWDD312 pKa = 3.3VGTDD316 pKa = 3.68SSGQAYY322 pKa = 10.42NSQFVNDD329 pKa = 4.24YY330 pKa = 8.8GTFIQSQSIPSLDD343 pKa = 3.27GAPILRR349 pKa = 11.84GVADD353 pKa = 3.27IRR355 pKa = 11.84IAQGSSFDD363 pKa = 3.85EE364 pKa = 4.3LSGVSAVDD372 pKa = 3.91NEE374 pKa = 4.79DD375 pKa = 3.35GDD377 pKa = 4.04LTAQIVLTGTVNTQVIDD394 pKa = 4.45DD395 pKa = 3.91YY396 pKa = 11.11TLTYY400 pKa = 10.21TVTDD404 pKa = 3.26SDD406 pKa = 4.54GNQATQARR414 pKa = 11.84VVTVFNNPPTLSGITDD430 pKa = 3.58TAIQTNDD437 pKa = 3.33NFDD440 pKa = 3.79ALTGVTASDD449 pKa = 4.56AEE451 pKa = 4.68DD452 pKa = 4.43GDD454 pKa = 4.23LTSQITVSGNVDD466 pKa = 3.03NSVEE470 pKa = 4.06GQYY473 pKa = 11.51VLTYY477 pKa = 10.08QVSDD481 pKa = 3.66SNGQSVTEE489 pKa = 3.93QRR491 pKa = 11.84TVTVSDD497 pKa = 3.66TPVNCTTEE505 pKa = 3.79WDD507 pKa = 3.64SNAIYY512 pKa = 8.89TQDD515 pKa = 4.69DD516 pKa = 3.65LADD519 pKa = 3.83YY520 pKa = 11.43DD521 pKa = 4.04GMTWRR526 pKa = 11.84AKK528 pKa = 8.34WWTQGNTPSDD538 pKa = 3.98DD539 pKa = 3.96PNGPWEE545 pKa = 4.0LTGDD549 pKa = 4.11SGCTTPNPDD558 pKa = 3.98PTPDD562 pKa = 4.57PDD564 pKa = 3.88PTPDD568 pKa = 4.54PDD570 pKa = 3.63PTPDD574 pKa = 3.37QGNGNYY580 pKa = 10.55ANYY583 pKa = 11.0AEE585 pKa = 4.34GTNYY589 pKa = 9.87QAGDD593 pKa = 3.53RR594 pKa = 11.84VSHH597 pKa = 5.94QGQDD601 pKa = 4.09FEE603 pKa = 5.38CKK605 pKa = 9.82PFPFSGWCSGSASAYY620 pKa = 10.39APNTGTNWQDD630 pKa = 2.25AWIRR634 pKa = 11.84LL635 pKa = 3.79
MM1 pKa = 6.48TTLSIGLIMAFQSQADD17 pKa = 3.74QTQMVDD23 pKa = 3.08QPGSVLVGYY32 pKa = 7.51WHH34 pKa = 6.7NFCNADD40 pKa = 3.35GYY42 pKa = 11.21QRR44 pKa = 11.84GNAPCISLTSTNPLYY59 pKa = 11.1NVVNVSFMRR68 pKa = 11.84VNDD71 pKa = 3.82TSQGRR76 pKa = 11.84IPTFEE81 pKa = 5.09LDD83 pKa = 3.26PGAGLSEE90 pKa = 4.06QEE92 pKa = 4.93FINQIGQLNAQGRR105 pKa = 11.84SVLIALGGADD115 pKa = 3.95AHH117 pKa = 7.34IEE119 pKa = 4.04LQTGDD124 pKa = 3.59EE125 pKa = 4.0QAFADD130 pKa = 4.53EE131 pKa = 4.98IIRR134 pKa = 11.84LVEE137 pKa = 4.05TYY139 pKa = 10.83GFDD142 pKa = 4.08GLDD145 pKa = 3.15IDD147 pKa = 5.55LEE149 pKa = 4.31QAAITARR156 pKa = 11.84DD157 pKa = 3.8NQTVIPAALRR167 pKa = 11.84MVKK170 pKa = 10.09DD171 pKa = 3.78HH172 pKa = 6.74YY173 pKa = 10.58RR174 pKa = 11.84AQGKK178 pKa = 9.06NFMITMAPEE187 pKa = 4.17FPYY190 pKa = 9.93LTGAGAYY197 pKa = 9.57VPYY200 pKa = 10.58LQGLEE205 pKa = 4.24GYY207 pKa = 10.35YY208 pKa = 10.88DD209 pKa = 4.57FINPQFYY216 pKa = 10.67NQGGDD221 pKa = 3.18GLYY224 pKa = 9.73IDD226 pKa = 4.17GVGWIAQNNDD236 pKa = 2.95ALKK239 pKa = 10.84EE240 pKa = 3.9EE241 pKa = 4.81FIYY244 pKa = 10.84YY245 pKa = 10.05ISDD248 pKa = 4.59AIANGSRR255 pKa = 11.84GFAQIPAEE263 pKa = 3.8RR264 pKa = 11.84LVFGIPANNDD274 pKa = 2.72AAATGFVADD283 pKa = 4.15PADD286 pKa = 4.28LYY288 pKa = 11.09RR289 pKa = 11.84AFGRR293 pKa = 11.84MRR295 pKa = 11.84EE296 pKa = 4.01QGQPLRR302 pKa = 11.84GIMTWSINWDD312 pKa = 3.3VGTDD316 pKa = 3.68SSGQAYY322 pKa = 10.42NSQFVNDD329 pKa = 4.24YY330 pKa = 8.8GTFIQSQSIPSLDD343 pKa = 3.27GAPILRR349 pKa = 11.84GVADD353 pKa = 3.27IRR355 pKa = 11.84IAQGSSFDD363 pKa = 3.85EE364 pKa = 4.3LSGVSAVDD372 pKa = 3.91NEE374 pKa = 4.79DD375 pKa = 3.35GDD377 pKa = 4.04LTAQIVLTGTVNTQVIDD394 pKa = 4.45DD395 pKa = 3.91YY396 pKa = 11.11TLTYY400 pKa = 10.21TVTDD404 pKa = 3.26SDD406 pKa = 4.54GNQATQARR414 pKa = 11.84VVTVFNNPPTLSGITDD430 pKa = 3.58TAIQTNDD437 pKa = 3.33NFDD440 pKa = 3.79ALTGVTASDD449 pKa = 4.56AEE451 pKa = 4.68DD452 pKa = 4.43GDD454 pKa = 4.23LTSQITVSGNVDD466 pKa = 3.03NSVEE470 pKa = 4.06GQYY473 pKa = 11.51VLTYY477 pKa = 10.08QVSDD481 pKa = 3.66SNGQSVTEE489 pKa = 3.93QRR491 pKa = 11.84TVTVSDD497 pKa = 3.66TPVNCTTEE505 pKa = 3.79WDD507 pKa = 3.64SNAIYY512 pKa = 8.89TQDD515 pKa = 4.69DD516 pKa = 3.65LADD519 pKa = 3.83YY520 pKa = 11.43DD521 pKa = 4.04GMTWRR526 pKa = 11.84AKK528 pKa = 8.34WWTQGNTPSDD538 pKa = 3.98DD539 pKa = 3.96PNGPWEE545 pKa = 4.0LTGDD549 pKa = 4.11SGCTTPNPDD558 pKa = 3.98PTPDD562 pKa = 4.57PDD564 pKa = 3.88PTPDD568 pKa = 4.54PDD570 pKa = 3.63PTPDD574 pKa = 3.37QGNGNYY580 pKa = 10.55ANYY583 pKa = 11.0AEE585 pKa = 4.34GTNYY589 pKa = 9.87QAGDD593 pKa = 3.53RR594 pKa = 11.84VSHH597 pKa = 5.94QGQDD601 pKa = 4.09FEE603 pKa = 5.38CKK605 pKa = 9.82PFPFSGWCSGSASAYY620 pKa = 10.39APNTGTNWQDD630 pKa = 2.25AWIRR634 pKa = 11.84LL635 pKa = 3.79
Molecular weight: 68.77 kDa
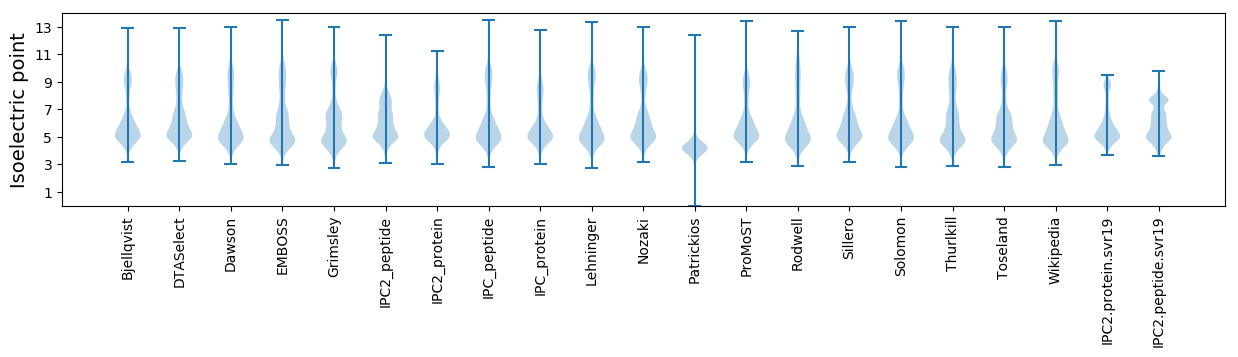
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R5H075|A0A4R5H075_9ALTE Protein Smg homolog OS=Alteromonadaceae bacterium M269 OX=2546219 GN=smg PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNIKK11 pKa = 10.11RR12 pKa = 11.84KK13 pKa = 9.04RR14 pKa = 11.84SHH16 pKa = 6.18GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 9.2VLASRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.27GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNIKK11 pKa = 10.11RR12 pKa = 11.84KK13 pKa = 9.04RR14 pKa = 11.84SHH16 pKa = 6.18GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 9.2VLASRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.27GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
Molecular weight: 5.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1643643 |
25 |
9140 |
337.4 |
37.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.989 ± 0.034 | 0.937 ± 0.012 |
5.847 ± 0.036 | 6.562 ± 0.046 |
4.405 ± 0.027 | 6.64 ± 0.035 |
2.108 ± 0.018 | 6.413 ± 0.027 |
5.376 ± 0.04 | 10.161 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.288 ± 0.017 | 4.706 ± 0.027 |
3.854 ± 0.022 | 4.522 ± 0.025 |
4.547 ± 0.021 | 7.12 ± 0.03 |
5.384 ± 0.03 | 6.795 ± 0.025 |
1.243 ± 0.014 | 3.104 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
