
Rice yellow stunt virus (RYSV) (Rice transitory yellowing virus)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Nucleorhabdovirus
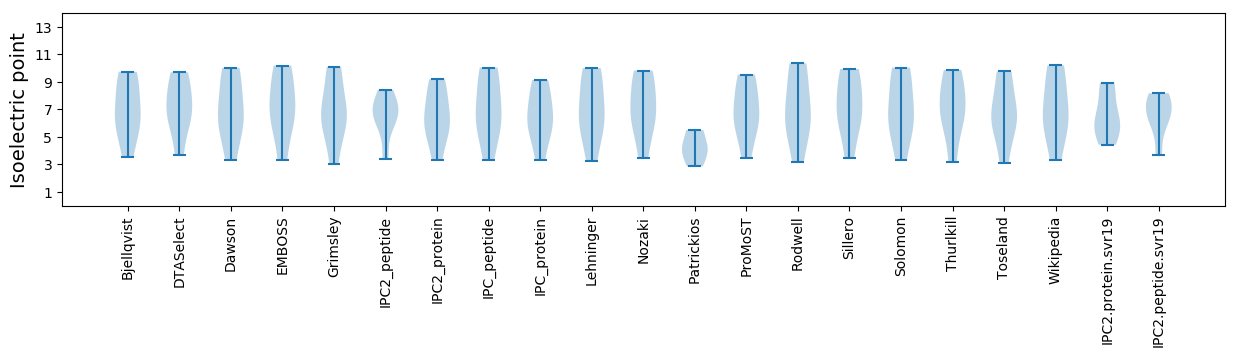
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>sp|Q86523|NCAP_RYSV Nucleoprotein OS=Rice yellow stunt virus OX=59380 GN=N PE=1 SV=1
MM1 pKa = 7.95SSQQEE6 pKa = 4.46TNDD9 pKa = 3.16KK10 pKa = 11.25SNTQGHH16 pKa = 7.27PEE18 pKa = 3.93TDD20 pKa = 3.56PEE22 pKa = 4.7GKK24 pKa = 8.72TGTDD28 pKa = 2.85TGNTEE33 pKa = 4.86DD34 pKa = 5.37SPPDD38 pKa = 3.56TDD40 pKa = 4.29NVPITDD46 pKa = 4.16DD47 pKa = 4.87AIMDD51 pKa = 3.94DD52 pKa = 5.39VMDD55 pKa = 4.63EE56 pKa = 4.15DD57 pKa = 4.81VKK59 pKa = 11.5EE60 pKa = 3.89EE61 pKa = 4.88DD62 pKa = 4.46IDD64 pKa = 4.15YY65 pKa = 11.23SWIEE69 pKa = 3.99DD70 pKa = 3.64MRR72 pKa = 11.84DD73 pKa = 3.14EE74 pKa = 5.4DD75 pKa = 6.05VDD77 pKa = 4.31AEE79 pKa = 4.29WLFEE83 pKa = 6.64LIDD86 pKa = 4.21EE87 pKa = 4.74CNGWPDD93 pKa = 2.93
MM1 pKa = 7.95SSQQEE6 pKa = 4.46TNDD9 pKa = 3.16KK10 pKa = 11.25SNTQGHH16 pKa = 7.27PEE18 pKa = 3.93TDD20 pKa = 3.56PEE22 pKa = 4.7GKK24 pKa = 8.72TGTDD28 pKa = 2.85TGNTEE33 pKa = 4.86DD34 pKa = 5.37SPPDD38 pKa = 3.56TDD40 pKa = 4.29NVPITDD46 pKa = 4.16DD47 pKa = 4.87AIMDD51 pKa = 3.94DD52 pKa = 5.39VMDD55 pKa = 4.63EE56 pKa = 4.15DD57 pKa = 4.81VKK59 pKa = 11.5EE60 pKa = 3.89EE61 pKa = 4.88DD62 pKa = 4.46IDD64 pKa = 4.15YY65 pKa = 11.23SWIEE69 pKa = 3.99DD70 pKa = 3.64MRR72 pKa = 11.84DD73 pKa = 3.14EE74 pKa = 5.4DD75 pKa = 6.05VDD77 pKa = 4.31AEE79 pKa = 4.29WLFEE83 pKa = 6.64LIDD86 pKa = 4.21EE87 pKa = 4.74CNGWPDD93 pKa = 2.93
Molecular weight: 10.54 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q98663|MVP_RYSV Movement protein OS=Rice yellow stunt virus OX=59380 GN=3 PE=1 SV=1
MM1 pKa = 7.54ANDD4 pKa = 3.63NVSDD8 pKa = 4.09YY9 pKa = 12.02ANAAPFARR17 pKa = 11.84FANLQNRR24 pKa = 11.84EE25 pKa = 4.06TLNPIGNEE33 pKa = 3.81AKK35 pKa = 10.16EE36 pKa = 3.9IPYY39 pKa = 10.49NRR41 pKa = 11.84DD42 pKa = 2.87QYY44 pKa = 10.44LTWLAEE50 pKa = 4.06GKK52 pKa = 10.04LFQIGALTDD61 pKa = 3.73AEE63 pKa = 5.05IVAAWTTIKK72 pKa = 9.84TAMGNNTFSEE82 pKa = 4.06THH84 pKa = 5.47MRR86 pKa = 11.84SIVKK90 pKa = 9.66IACNLRR96 pKa = 11.84GITPGSTPLLVTYY109 pKa = 9.58NPPQSATWAPAPSTDD124 pKa = 4.27AIYY127 pKa = 10.82SGTPVAGVIIPQNTGAGGEE146 pKa = 4.17DD147 pKa = 4.07TEE149 pKa = 4.8TEE151 pKa = 3.87ASKK154 pKa = 11.09ARR156 pKa = 11.84AIAFICCYY164 pKa = 10.04LLRR167 pKa = 11.84FIVKK171 pKa = 9.05TEE173 pKa = 3.73EE174 pKa = 4.0HH175 pKa = 6.15LTNSLGNLKK184 pKa = 10.04LQYY187 pKa = 10.5SRR189 pKa = 11.84LYY191 pKa = 9.64SAQSATLSNWNPSNTWASRR210 pKa = 11.84VKK212 pKa = 10.79LGFDD216 pKa = 3.24TYY218 pKa = 10.07LTLRR222 pKa = 11.84ATVAYY227 pKa = 10.08NIASADD233 pKa = 3.5ALLVPEE239 pKa = 4.13NVNYY243 pKa = 10.08GLCRR247 pKa = 11.84MLVFQHH253 pKa = 6.73LEE255 pKa = 3.81LSGLQLYY262 pKa = 9.32KK263 pKa = 10.13MAMTLIAHH271 pKa = 6.89FKK273 pKa = 10.48LIEE276 pKa = 4.36PNKK279 pKa = 8.64FLSWIYY285 pKa = 11.05DD286 pKa = 3.8PLSEE290 pKa = 4.96ASIDD294 pKa = 3.73QIYY297 pKa = 9.97KK298 pKa = 9.96IAVNYY303 pKa = 10.89DD304 pKa = 3.13NVNSKK309 pKa = 6.97THH311 pKa = 5.52KK312 pKa = 7.6HH313 pKa = 3.99WKK315 pKa = 7.39YY316 pKa = 10.75AKK318 pKa = 10.06LARR321 pKa = 11.84GQYY324 pKa = 8.37WLNTTVKK331 pKa = 10.09RR332 pKa = 11.84NQFLAYY338 pKa = 9.52ILADD342 pKa = 3.8LEE344 pKa = 4.58LKK346 pKa = 10.78YY347 pKa = 11.14GLAGKK352 pKa = 9.8SDD354 pKa = 3.66YY355 pKa = 11.23SSPKK359 pKa = 9.82RR360 pKa = 11.84MKK362 pKa = 10.02ALSGMPVEE370 pKa = 5.07RR371 pKa = 11.84MTEE374 pKa = 3.64AEE376 pKa = 4.47TISKK380 pKa = 10.27AVEE383 pKa = 3.81QMYY386 pKa = 8.57TAIEE390 pKa = 3.98SAKK393 pKa = 10.55RR394 pKa = 11.84VDD396 pKa = 3.27AGAAYY401 pKa = 10.37RR402 pKa = 11.84LAKK405 pKa = 10.54KK406 pKa = 10.46LGPPRR411 pKa = 11.84ANAHH415 pKa = 5.36SRR417 pKa = 11.84RR418 pKa = 11.84KK419 pKa = 9.5EE420 pKa = 3.95PNNSRR425 pKa = 11.84QHH427 pKa = 6.76RR428 pKa = 11.84DD429 pKa = 3.03KK430 pKa = 11.24QPNSKK435 pKa = 9.37QQGRR439 pKa = 11.84DD440 pKa = 3.04KK441 pKa = 10.99RR442 pKa = 11.84NKK444 pKa = 8.05HH445 pKa = 4.66QVLGRR450 pKa = 11.84HH451 pKa = 6.17SKK453 pKa = 9.79PQAQGPPNKK462 pKa = 9.94QQDD465 pKa = 4.09LGPHH469 pKa = 5.08NNKK472 pKa = 9.54PKK474 pKa = 9.86EE475 pKa = 4.03ASRR478 pKa = 11.84PPQQDD483 pKa = 3.1RR484 pKa = 11.84QQLAQPWRR492 pKa = 11.84LTRR495 pKa = 11.84RR496 pKa = 11.84QRR498 pKa = 11.84GARR501 pKa = 11.84GHH503 pKa = 5.69RR504 pKa = 11.84TQTLSGMFCKK514 pKa = 10.1GTCQCIQQ521 pKa = 2.97
MM1 pKa = 7.54ANDD4 pKa = 3.63NVSDD8 pKa = 4.09YY9 pKa = 12.02ANAAPFARR17 pKa = 11.84FANLQNRR24 pKa = 11.84EE25 pKa = 4.06TLNPIGNEE33 pKa = 3.81AKK35 pKa = 10.16EE36 pKa = 3.9IPYY39 pKa = 10.49NRR41 pKa = 11.84DD42 pKa = 2.87QYY44 pKa = 10.44LTWLAEE50 pKa = 4.06GKK52 pKa = 10.04LFQIGALTDD61 pKa = 3.73AEE63 pKa = 5.05IVAAWTTIKK72 pKa = 9.84TAMGNNTFSEE82 pKa = 4.06THH84 pKa = 5.47MRR86 pKa = 11.84SIVKK90 pKa = 9.66IACNLRR96 pKa = 11.84GITPGSTPLLVTYY109 pKa = 9.58NPPQSATWAPAPSTDD124 pKa = 4.27AIYY127 pKa = 10.82SGTPVAGVIIPQNTGAGGEE146 pKa = 4.17DD147 pKa = 4.07TEE149 pKa = 4.8TEE151 pKa = 3.87ASKK154 pKa = 11.09ARR156 pKa = 11.84AIAFICCYY164 pKa = 10.04LLRR167 pKa = 11.84FIVKK171 pKa = 9.05TEE173 pKa = 3.73EE174 pKa = 4.0HH175 pKa = 6.15LTNSLGNLKK184 pKa = 10.04LQYY187 pKa = 10.5SRR189 pKa = 11.84LYY191 pKa = 9.64SAQSATLSNWNPSNTWASRR210 pKa = 11.84VKK212 pKa = 10.79LGFDD216 pKa = 3.24TYY218 pKa = 10.07LTLRR222 pKa = 11.84ATVAYY227 pKa = 10.08NIASADD233 pKa = 3.5ALLVPEE239 pKa = 4.13NVNYY243 pKa = 10.08GLCRR247 pKa = 11.84MLVFQHH253 pKa = 6.73LEE255 pKa = 3.81LSGLQLYY262 pKa = 9.32KK263 pKa = 10.13MAMTLIAHH271 pKa = 6.89FKK273 pKa = 10.48LIEE276 pKa = 4.36PNKK279 pKa = 8.64FLSWIYY285 pKa = 11.05DD286 pKa = 3.8PLSEE290 pKa = 4.96ASIDD294 pKa = 3.73QIYY297 pKa = 9.97KK298 pKa = 9.96IAVNYY303 pKa = 10.89DD304 pKa = 3.13NVNSKK309 pKa = 6.97THH311 pKa = 5.52KK312 pKa = 7.6HH313 pKa = 3.99WKK315 pKa = 7.39YY316 pKa = 10.75AKK318 pKa = 10.06LARR321 pKa = 11.84GQYY324 pKa = 8.37WLNTTVKK331 pKa = 10.09RR332 pKa = 11.84NQFLAYY338 pKa = 9.52ILADD342 pKa = 3.8LEE344 pKa = 4.58LKK346 pKa = 10.78YY347 pKa = 11.14GLAGKK352 pKa = 9.8SDD354 pKa = 3.66YY355 pKa = 11.23SSPKK359 pKa = 9.82RR360 pKa = 11.84MKK362 pKa = 10.02ALSGMPVEE370 pKa = 5.07RR371 pKa = 11.84MTEE374 pKa = 3.64AEE376 pKa = 4.47TISKK380 pKa = 10.27AVEE383 pKa = 3.81QMYY386 pKa = 8.57TAIEE390 pKa = 3.98SAKK393 pKa = 10.55RR394 pKa = 11.84VDD396 pKa = 3.27AGAAYY401 pKa = 10.37RR402 pKa = 11.84LAKK405 pKa = 10.54KK406 pKa = 10.46LGPPRR411 pKa = 11.84ANAHH415 pKa = 5.36SRR417 pKa = 11.84RR418 pKa = 11.84KK419 pKa = 9.5EE420 pKa = 3.95PNNSRR425 pKa = 11.84QHH427 pKa = 6.76RR428 pKa = 11.84DD429 pKa = 3.03KK430 pKa = 11.24QPNSKK435 pKa = 9.37QQGRR439 pKa = 11.84DD440 pKa = 3.04KK441 pKa = 10.99RR442 pKa = 11.84NKK444 pKa = 8.05HH445 pKa = 4.66QVLGRR450 pKa = 11.84HH451 pKa = 6.17SKK453 pKa = 9.79PQAQGPPNKK462 pKa = 9.94QQDD465 pKa = 4.09LGPHH469 pKa = 5.08NNKK472 pKa = 9.54PKK474 pKa = 9.86EE475 pKa = 4.03ASRR478 pKa = 11.84PPQQDD483 pKa = 3.1RR484 pKa = 11.84QQLAQPWRR492 pKa = 11.84LTRR495 pKa = 11.84RR496 pKa = 11.84QRR498 pKa = 11.84GARR501 pKa = 11.84GHH503 pKa = 5.69RR504 pKa = 11.84TQTLSGMFCKK514 pKa = 10.1GTCQCIQQ521 pKa = 2.97
Molecular weight: 58.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4128 |
93 |
1967 |
589.7 |
66.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.281 ± 0.954 | 1.817 ± 0.247 |
6.105 ± 0.605 | 5.644 ± 0.406 |
2.98 ± 0.369 | 6.032 ± 0.212 |
2.398 ± 0.342 | 7.461 ± 0.546 |
6.202 ± 0.521 | 8.963 ± 0.714 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.004 ± 0.234 | 4.918 ± 0.504 |
4.385 ± 0.353 | 3.319 ± 0.494 |
4.869 ± 0.394 | 8.067 ± 0.575 |
6.977 ± 0.473 | 6.008 ± 0.481 |
1.55 ± 0.163 | 4.021 ± 0.368 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
