
Siphoviridae sp. ctCJE6
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.47
Get precalculated fractions of proteins
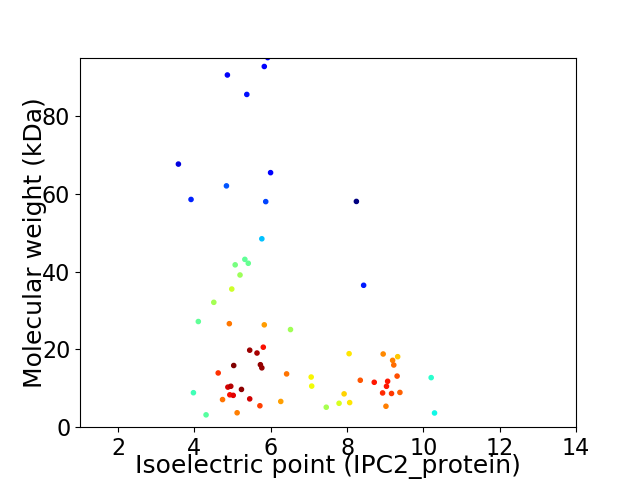
Virtual 2D-PAGE plot for 64 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
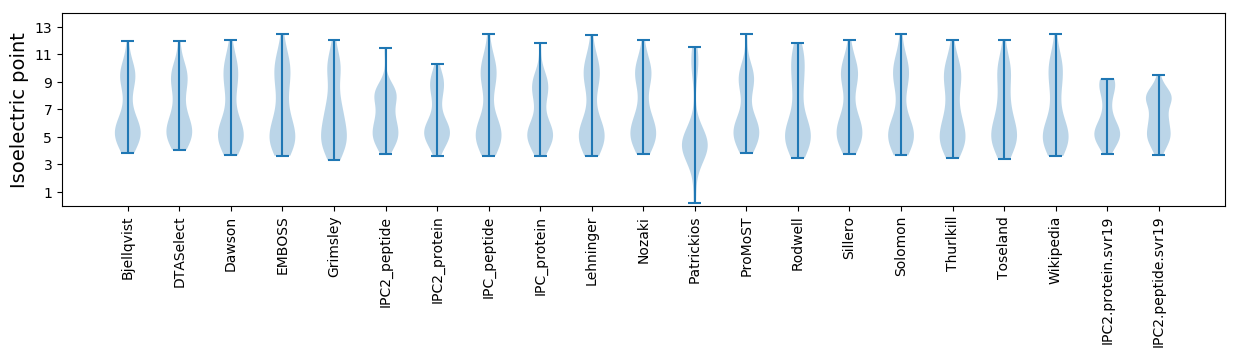
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5Q2W9Q8|A0A5Q2W9Q8_9CAUD Capsid maturation protease OS=Siphoviridae sp. ctCJE6 OX=2656719 PE=4 SV=1
MM1 pKa = 7.75RR2 pKa = 11.84FKK4 pKa = 11.14LLRR7 pKa = 11.84DD8 pKa = 3.29SFVFVFLMTFASLFMNLAIVGQKK31 pKa = 10.65VMGQPAPGPVLDD43 pKa = 3.58MAEE46 pKa = 5.25FIRR49 pKa = 11.84MDD51 pKa = 3.34TTNFNGHH58 pKa = 7.08LSSADD63 pKa = 3.31SDD65 pKa = 3.98VQKK68 pKa = 11.13ALEE71 pKa = 4.17TLDD74 pKa = 4.58DD75 pKa = 4.36LVAGGHH81 pKa = 6.27GNGANCSAGNFPLGVDD97 pKa = 3.31EE98 pKa = 4.93NGAVEE103 pKa = 4.49SCTDD107 pKa = 2.77AWTEE111 pKa = 4.38AEE113 pKa = 4.19NTSAGYY119 pKa = 10.49YY120 pKa = 10.3NSLSDD125 pKa = 4.05LQTAVSNDD133 pKa = 2.89FHH135 pKa = 8.09NLGGTDD141 pKa = 4.34DD142 pKa = 5.21DD143 pKa = 4.93VPDD146 pKa = 4.36SGDD149 pKa = 3.38LGLIDD154 pKa = 3.92TEE156 pKa = 4.92AEE158 pKa = 4.13FEE160 pKa = 4.5SEE162 pKa = 4.44LFPIFTPDD170 pKa = 5.18DD171 pKa = 3.86GALDD175 pKa = 4.99DD176 pKa = 6.78DD177 pKa = 6.42DD178 pKa = 4.4ITDD181 pKa = 4.26DD182 pKa = 4.43ASTSLTDD189 pKa = 3.43TGNIAYY195 pKa = 10.1LNSAEE200 pKa = 4.48NISGVWEE207 pKa = 4.16IQDD210 pKa = 3.46DD211 pKa = 4.06TLLNFGNSADD221 pKa = 3.83GGIQHH226 pKa = 6.31VSSSTTTRR234 pKa = 11.84YY235 pKa = 10.41SGVPFHH241 pKa = 7.01FGADD245 pKa = 3.41GTGADD250 pKa = 3.74VVFNSDD256 pKa = 2.7TSGAQLTYY264 pKa = 10.87DD265 pKa = 4.47PSPANATTGHH275 pKa = 7.92LIQDD279 pKa = 4.26VYY281 pKa = 10.51NGQMVYY287 pKa = 9.79WEE289 pKa = 4.56LSGSNNSTSTSSKK302 pKa = 8.9GHH304 pKa = 6.46LNIDD308 pKa = 3.21VDD310 pKa = 4.35FTGSYY315 pKa = 10.93SSLQGANALYY325 pKa = 10.51FDD327 pKa = 4.64FADD330 pKa = 3.74ARR332 pKa = 11.84NISGSFGATGTRR344 pKa = 11.84IASGKK349 pKa = 8.48IARR352 pKa = 11.84TGTHH356 pKa = 6.06SGSSTAEE363 pKa = 4.02STQMLYY369 pKa = 10.06MDD371 pKa = 4.99MGDD374 pKa = 4.18GMTLSGTPTVSRR386 pKa = 11.84TFADD390 pKa = 3.96FLGTSIQGVSSTGATSAVHH409 pKa = 6.56RR410 pKa = 11.84GVHH413 pKa = 5.47LHH415 pKa = 5.93NGYY418 pKa = 9.85IVFATGGGTHH428 pKa = 6.43TFSHH432 pKa = 6.45YY433 pKa = 11.04GVDD436 pKa = 3.13IANTFSEE443 pKa = 4.89VEE445 pKa = 3.87VSGTVNGTNIGVWYY459 pKa = 9.22HH460 pKa = 5.96PTMSSTPVAEE470 pKa = 4.51YY471 pKa = 10.92AFKK474 pKa = 10.8ADD476 pKa = 3.85RR477 pKa = 11.84SIIALKK483 pKa = 10.66SDD485 pKa = 3.48GSTGAYY491 pKa = 9.63AGGRR495 pKa = 11.84AVFGAGEE502 pKa = 4.06DD503 pKa = 3.54AEE505 pKa = 4.4IYY507 pKa = 10.89YY508 pKa = 10.73DD509 pKa = 3.91GTDD512 pKa = 4.36LIIDD516 pKa = 4.22AKK518 pKa = 10.82LVGSGAVDD526 pKa = 3.0IEE528 pKa = 4.65SEE530 pKa = 5.24LEE532 pKa = 3.92VDD534 pKa = 5.09DD535 pKa = 6.32LNTGGNAGTDD545 pKa = 3.43LCHH548 pKa = 7.1DD549 pKa = 4.08ANGRR553 pKa = 11.84LCACGSCAA561 pKa = 5.03
MM1 pKa = 7.75RR2 pKa = 11.84FKK4 pKa = 11.14LLRR7 pKa = 11.84DD8 pKa = 3.29SFVFVFLMTFASLFMNLAIVGQKK31 pKa = 10.65VMGQPAPGPVLDD43 pKa = 3.58MAEE46 pKa = 5.25FIRR49 pKa = 11.84MDD51 pKa = 3.34TTNFNGHH58 pKa = 7.08LSSADD63 pKa = 3.31SDD65 pKa = 3.98VQKK68 pKa = 11.13ALEE71 pKa = 4.17TLDD74 pKa = 4.58DD75 pKa = 4.36LVAGGHH81 pKa = 6.27GNGANCSAGNFPLGVDD97 pKa = 3.31EE98 pKa = 4.93NGAVEE103 pKa = 4.49SCTDD107 pKa = 2.77AWTEE111 pKa = 4.38AEE113 pKa = 4.19NTSAGYY119 pKa = 10.49YY120 pKa = 10.3NSLSDD125 pKa = 4.05LQTAVSNDD133 pKa = 2.89FHH135 pKa = 8.09NLGGTDD141 pKa = 4.34DD142 pKa = 5.21DD143 pKa = 4.93VPDD146 pKa = 4.36SGDD149 pKa = 3.38LGLIDD154 pKa = 3.92TEE156 pKa = 4.92AEE158 pKa = 4.13FEE160 pKa = 4.5SEE162 pKa = 4.44LFPIFTPDD170 pKa = 5.18DD171 pKa = 3.86GALDD175 pKa = 4.99DD176 pKa = 6.78DD177 pKa = 6.42DD178 pKa = 4.4ITDD181 pKa = 4.26DD182 pKa = 4.43ASTSLTDD189 pKa = 3.43TGNIAYY195 pKa = 10.1LNSAEE200 pKa = 4.48NISGVWEE207 pKa = 4.16IQDD210 pKa = 3.46DD211 pKa = 4.06TLLNFGNSADD221 pKa = 3.83GGIQHH226 pKa = 6.31VSSSTTTRR234 pKa = 11.84YY235 pKa = 10.41SGVPFHH241 pKa = 7.01FGADD245 pKa = 3.41GTGADD250 pKa = 3.74VVFNSDD256 pKa = 2.7TSGAQLTYY264 pKa = 10.87DD265 pKa = 4.47PSPANATTGHH275 pKa = 7.92LIQDD279 pKa = 4.26VYY281 pKa = 10.51NGQMVYY287 pKa = 9.79WEE289 pKa = 4.56LSGSNNSTSTSSKK302 pKa = 8.9GHH304 pKa = 6.46LNIDD308 pKa = 3.21VDD310 pKa = 4.35FTGSYY315 pKa = 10.93SSLQGANALYY325 pKa = 10.51FDD327 pKa = 4.64FADD330 pKa = 3.74ARR332 pKa = 11.84NISGSFGATGTRR344 pKa = 11.84IASGKK349 pKa = 8.48IARR352 pKa = 11.84TGTHH356 pKa = 6.06SGSSTAEE363 pKa = 4.02STQMLYY369 pKa = 10.06MDD371 pKa = 4.99MGDD374 pKa = 4.18GMTLSGTPTVSRR386 pKa = 11.84TFADD390 pKa = 3.96FLGTSIQGVSSTGATSAVHH409 pKa = 6.56RR410 pKa = 11.84GVHH413 pKa = 5.47LHH415 pKa = 5.93NGYY418 pKa = 9.85IVFATGGGTHH428 pKa = 6.43TFSHH432 pKa = 6.45YY433 pKa = 11.04GVDD436 pKa = 3.13IANTFSEE443 pKa = 4.89VEE445 pKa = 3.87VSGTVNGTNIGVWYY459 pKa = 9.22HH460 pKa = 5.96PTMSSTPVAEE470 pKa = 4.51YY471 pKa = 10.92AFKK474 pKa = 10.8ADD476 pKa = 3.85RR477 pKa = 11.84SIIALKK483 pKa = 10.66SDD485 pKa = 3.48GSTGAYY491 pKa = 9.63AGGRR495 pKa = 11.84AVFGAGEE502 pKa = 4.06DD503 pKa = 3.54AEE505 pKa = 4.4IYY507 pKa = 10.89YY508 pKa = 10.73DD509 pKa = 3.91GTDD512 pKa = 4.36LIIDD516 pKa = 4.22AKK518 pKa = 10.82LVGSGAVDD526 pKa = 3.0IEE528 pKa = 4.65SEE530 pKa = 5.24LEE532 pKa = 3.92VDD534 pKa = 5.09DD535 pKa = 6.32LNTGGNAGTDD545 pKa = 3.43LCHH548 pKa = 7.1DD549 pKa = 4.08ANGRR553 pKa = 11.84LCACGSCAA561 pKa = 5.03
Molecular weight: 58.54 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5Q2W9C2|A0A5Q2W9C2_9CAUD Uncharacterized protein OS=Siphoviridae sp. ctCJE6 OX=2656719 PE=4 SV=1
MM1 pKa = 8.01RR2 pKa = 11.84LTKK5 pKa = 10.44NDD7 pKa = 2.98WDD9 pKa = 3.23ILMFYY14 pKa = 10.62LRR16 pKa = 11.84AAVVANVILFSCVLMAHH33 pKa = 6.9AEE35 pKa = 4.46SVQPPHH41 pKa = 6.56NLWKK45 pKa = 10.77GLIAEE50 pKa = 4.7AVSEE54 pKa = 4.51GPEE57 pKa = 4.0GMVAVCLVYY66 pKa = 10.53RR67 pKa = 11.84NRR69 pKa = 11.84LQAGMRR75 pKa = 11.84MGCSGLKK82 pKa = 10.17RR83 pKa = 11.84RR84 pKa = 11.84DD85 pKa = 2.98LDD87 pKa = 3.54QFVARR92 pKa = 11.84QGSRR96 pKa = 11.84YY97 pKa = 8.97EE98 pKa = 4.58RR99 pKa = 11.84MAKK102 pKa = 10.35DD103 pKa = 2.87IVARR107 pKa = 11.84VFSGRR112 pKa = 11.84EE113 pKa = 3.35RR114 pKa = 11.84DD115 pKa = 3.59VTMGATHH122 pKa = 6.44YY123 pKa = 11.09EE124 pKa = 4.2NITAFGMPWWAKK136 pKa = 8.24SMRR139 pKa = 11.84ITARR143 pKa = 11.84IGSHH147 pKa = 5.25TFFKK151 pKa = 11.01
MM1 pKa = 8.01RR2 pKa = 11.84LTKK5 pKa = 10.44NDD7 pKa = 2.98WDD9 pKa = 3.23ILMFYY14 pKa = 10.62LRR16 pKa = 11.84AAVVANVILFSCVLMAHH33 pKa = 6.9AEE35 pKa = 4.46SVQPPHH41 pKa = 6.56NLWKK45 pKa = 10.77GLIAEE50 pKa = 4.7AVSEE54 pKa = 4.51GPEE57 pKa = 4.0GMVAVCLVYY66 pKa = 10.53RR67 pKa = 11.84NRR69 pKa = 11.84LQAGMRR75 pKa = 11.84MGCSGLKK82 pKa = 10.17RR83 pKa = 11.84RR84 pKa = 11.84DD85 pKa = 2.98LDD87 pKa = 3.54QFVARR92 pKa = 11.84QGSRR96 pKa = 11.84YY97 pKa = 8.97EE98 pKa = 4.58RR99 pKa = 11.84MAKK102 pKa = 10.35DD103 pKa = 2.87IVARR107 pKa = 11.84VFSGRR112 pKa = 11.84EE113 pKa = 3.35RR114 pKa = 11.84DD115 pKa = 3.59VTMGATHH122 pKa = 6.44YY123 pKa = 11.09EE124 pKa = 4.2NITAFGMPWWAKK136 pKa = 8.24SMRR139 pKa = 11.84ITARR143 pKa = 11.84IGSHH147 pKa = 5.25TFFKK151 pKa = 11.01
Molecular weight: 17.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
14486 |
29 |
833 |
226.3 |
25.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.538 ± 0.463 | 1.319 ± 0.157 |
6.71 ± 0.255 | 6.751 ± 0.424 |
4.687 ± 0.227 | 7.525 ± 0.32 |
1.712 ± 0.154 | 6.917 ± 0.304 |
6.689 ± 0.521 | 7.718 ± 0.256 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.651 ± 0.176 | 4.791 ± 0.172 |
3.403 ± 0.247 | 3.189 ± 0.149 |
5.412 ± 0.345 | 6.227 ± 0.329 |
5.164 ± 0.428 | 6.772 ± 0.259 |
1.595 ± 0.156 | 3.231 ± 0.223 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
