
Chlamydotis macqueenii (Macqueen s bustard)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Sarcopterygii; Dipnotetrapodomorpha; Tetrapoda; Amniota; Sauropsida; Sauria;
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
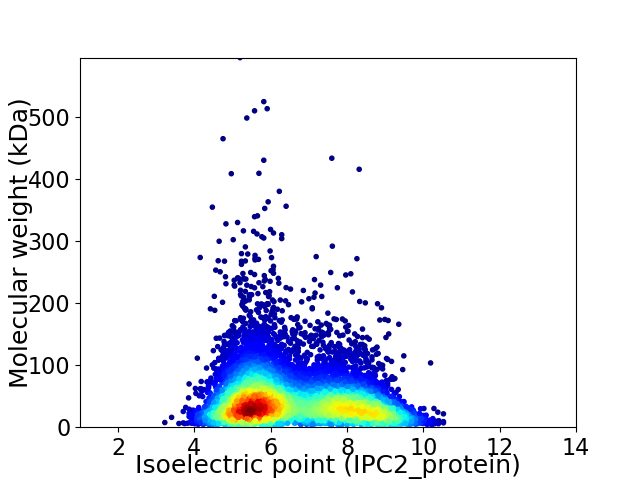
Virtual 2D-PAGE plot for 8277 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A091KLE7|A0A091KLE7_9GRUI Neuroblast differentiation-associated protein AHNAK (Fragment) OS=Chlamydotis macqueenii OX=187382 GN=N324_04926 PE=4 SV=1
QQ1 pKa = 7.23CRR3 pKa = 11.84YY4 pKa = 9.78SPCFPGVRR12 pKa = 11.84CVNTAPGFRR21 pKa = 11.84CEE23 pKa = 4.17TCPPGYY29 pKa = 9.16TGQTVQGVGLSYY41 pKa = 11.12AKK43 pKa = 10.53SNKK46 pKa = 7.95QVCLDD51 pKa = 3.73IDD53 pKa = 3.67EE54 pKa = 4.91CQNGGLGLCVPNSHH68 pKa = 7.46CINTLGSYY76 pKa = 10.2RR77 pKa = 11.84CGQCKK82 pKa = 9.57PGYY85 pKa = 9.17TGDD88 pKa = 3.61QARR91 pKa = 11.84GCQAEE96 pKa = 4.14RR97 pKa = 11.84SCRR100 pKa = 11.84NRR102 pKa = 11.84ALNPCSVHH110 pKa = 5.91ARR112 pKa = 11.84CIEE115 pKa = 4.02EE116 pKa = 3.8KK117 pKa = 10.27RR118 pKa = 11.84GEE120 pKa = 4.27VTCICGIGWAGDD132 pKa = 3.82GYY134 pKa = 10.69ICGKK138 pKa = 10.5DD139 pKa = 3.18IDD141 pKa = 4.02IDD143 pKa = 4.15GYY145 pKa = 10.37PNDD148 pKa = 4.44EE149 pKa = 4.52LSCSAEE155 pKa = 3.77NCRR158 pKa = 11.84KK159 pKa = 10.08DD160 pKa = 3.11NCRR163 pKa = 11.84FVPNSGQEE171 pKa = 4.02DD172 pKa = 4.12ADD174 pKa = 3.79GDD176 pKa = 4.61GIGDD180 pKa = 4.07ACDD183 pKa = 3.53EE184 pKa = 4.74DD185 pKa = 5.71ADD187 pKa = 4.46GDD189 pKa = 4.49GIPNEE194 pKa = 4.01QDD196 pKa = 3.11NCVLAPNVNQRR207 pKa = 11.84NVDD210 pKa = 3.19QDD212 pKa = 3.47IFGDD216 pKa = 3.6ACDD219 pKa = 3.4NCRR222 pKa = 11.84NILNNDD228 pKa = 3.09QRR230 pKa = 11.84DD231 pKa = 3.44TDD233 pKa = 3.87GDD235 pKa = 4.16GKK237 pKa = 11.42GDD239 pKa = 3.74ACDD242 pKa = 5.17DD243 pKa = 4.26DD244 pKa = 4.15MDD246 pKa = 6.26GDD248 pKa = 4.5GIKK251 pKa = 10.77NLLDD255 pKa = 3.41NCQRR259 pKa = 11.84IPNQDD264 pKa = 3.43QEE266 pKa = 5.62DD267 pKa = 3.92KK268 pKa = 11.46DD269 pKa = 3.9NDD271 pKa = 4.31GVGDD275 pKa = 4.14ACDD278 pKa = 3.51SCPTISNPNQSDD290 pKa = 3.32VDD292 pKa = 3.63NDD294 pKa = 4.07LVGDD298 pKa = 4.11SCDD301 pKa = 3.63TNQDD305 pKa = 3.04SDD307 pKa = 4.07GDD309 pKa = 4.03GHH311 pKa = 7.17QDD313 pKa = 3.22STDD316 pKa = 3.33NCPTIINSSQLDD328 pKa = 3.61TDD330 pKa = 3.69KK331 pKa = 11.62DD332 pKa = 4.04GLGDD336 pKa = 3.77EE337 pKa = 5.06CDD339 pKa = 4.04EE340 pKa = 6.11DD341 pKa = 5.22DD342 pKa = 5.9DD343 pKa = 5.67NDD345 pKa = 5.36GILDD349 pKa = 4.79LLPPGPDD356 pKa = 2.92NCRR359 pKa = 11.84LVPNPGQEE367 pKa = 4.18DD368 pKa = 4.25DD369 pKa = 4.42NGDD372 pKa = 3.69GVGDD376 pKa = 3.48ICEE379 pKa = 4.29SDD381 pKa = 3.45FDD383 pKa = 3.9QDD385 pKa = 3.89TVIDD389 pKa = 4.23QIDD392 pKa = 3.82VCPEE396 pKa = 3.54NAEE399 pKa = 3.78ITLTDD404 pKa = 3.83FRR406 pKa = 11.84AYY408 pKa = 8.19QTVVLDD414 pKa = 4.22PEE416 pKa = 5.32GDD418 pKa = 3.65AQIDD422 pKa = 4.13PNWVVLNQGMEE433 pKa = 4.11IVQTMNSDD441 pKa = 3.19PGLAVGYY448 pKa = 7.31TAFNGVDD455 pKa = 3.99FEE457 pKa = 4.97GTFHH461 pKa = 6.81VNTVTDD467 pKa = 3.57DD468 pKa = 4.08DD469 pKa = 4.35YY470 pKa = 12.0AGFIFGYY477 pKa = 9.58QDD479 pKa = 2.83SSSFYY484 pKa = 10.47VVMWKK489 pKa = 7.78QTEE492 pKa = 3.86QTYY495 pKa = 8.13WQATPFRR502 pKa = 11.84AVAEE506 pKa = 4.17PGIQLKK512 pKa = 10.07AVKK515 pKa = 10.09SKK517 pKa = 9.55TGPGEE522 pKa = 3.82HH523 pKa = 6.52LRR525 pKa = 11.84NSLWHH530 pKa = 6.51TGDD533 pKa = 3.06TSDD536 pKa = 5.19QVRR539 pKa = 11.84LLWKK543 pKa = 10.3DD544 pKa = 3.22PRR546 pKa = 11.84NVGWKK551 pKa = 10.39DD552 pKa = 3.18KK553 pKa = 11.21VSYY556 pKa = 10.33RR557 pKa = 11.84WFLQHH562 pKa = 6.9RR563 pKa = 11.84PQIGYY568 pKa = 9.07IRR570 pKa = 11.84ARR572 pKa = 11.84FYY574 pKa = 11.09EE575 pKa = 4.54GSDD578 pKa = 3.39LVADD582 pKa = 3.84SGVTIDD588 pKa = 3.09TTMRR592 pKa = 11.84GGRR595 pKa = 11.84LGVFCFSQEE604 pKa = 3.94NIIWSNLKK612 pKa = 9.68YY613 pKa = 10.31RR614 pKa = 11.84CNDD617 pKa = 4.18TIPEE621 pKa = 4.31DD622 pKa = 3.77FQEE625 pKa = 4.53FQAQQFGVGDD635 pKa = 3.49II636 pKa = 4.15
QQ1 pKa = 7.23CRR3 pKa = 11.84YY4 pKa = 9.78SPCFPGVRR12 pKa = 11.84CVNTAPGFRR21 pKa = 11.84CEE23 pKa = 4.17TCPPGYY29 pKa = 9.16TGQTVQGVGLSYY41 pKa = 11.12AKK43 pKa = 10.53SNKK46 pKa = 7.95QVCLDD51 pKa = 3.73IDD53 pKa = 3.67EE54 pKa = 4.91CQNGGLGLCVPNSHH68 pKa = 7.46CINTLGSYY76 pKa = 10.2RR77 pKa = 11.84CGQCKK82 pKa = 9.57PGYY85 pKa = 9.17TGDD88 pKa = 3.61QARR91 pKa = 11.84GCQAEE96 pKa = 4.14RR97 pKa = 11.84SCRR100 pKa = 11.84NRR102 pKa = 11.84ALNPCSVHH110 pKa = 5.91ARR112 pKa = 11.84CIEE115 pKa = 4.02EE116 pKa = 3.8KK117 pKa = 10.27RR118 pKa = 11.84GEE120 pKa = 4.27VTCICGIGWAGDD132 pKa = 3.82GYY134 pKa = 10.69ICGKK138 pKa = 10.5DD139 pKa = 3.18IDD141 pKa = 4.02IDD143 pKa = 4.15GYY145 pKa = 10.37PNDD148 pKa = 4.44EE149 pKa = 4.52LSCSAEE155 pKa = 3.77NCRR158 pKa = 11.84KK159 pKa = 10.08DD160 pKa = 3.11NCRR163 pKa = 11.84FVPNSGQEE171 pKa = 4.02DD172 pKa = 4.12ADD174 pKa = 3.79GDD176 pKa = 4.61GIGDD180 pKa = 4.07ACDD183 pKa = 3.53EE184 pKa = 4.74DD185 pKa = 5.71ADD187 pKa = 4.46GDD189 pKa = 4.49GIPNEE194 pKa = 4.01QDD196 pKa = 3.11NCVLAPNVNQRR207 pKa = 11.84NVDD210 pKa = 3.19QDD212 pKa = 3.47IFGDD216 pKa = 3.6ACDD219 pKa = 3.4NCRR222 pKa = 11.84NILNNDD228 pKa = 3.09QRR230 pKa = 11.84DD231 pKa = 3.44TDD233 pKa = 3.87GDD235 pKa = 4.16GKK237 pKa = 11.42GDD239 pKa = 3.74ACDD242 pKa = 5.17DD243 pKa = 4.26DD244 pKa = 4.15MDD246 pKa = 6.26GDD248 pKa = 4.5GIKK251 pKa = 10.77NLLDD255 pKa = 3.41NCQRR259 pKa = 11.84IPNQDD264 pKa = 3.43QEE266 pKa = 5.62DD267 pKa = 3.92KK268 pKa = 11.46DD269 pKa = 3.9NDD271 pKa = 4.31GVGDD275 pKa = 4.14ACDD278 pKa = 3.51SCPTISNPNQSDD290 pKa = 3.32VDD292 pKa = 3.63NDD294 pKa = 4.07LVGDD298 pKa = 4.11SCDD301 pKa = 3.63TNQDD305 pKa = 3.04SDD307 pKa = 4.07GDD309 pKa = 4.03GHH311 pKa = 7.17QDD313 pKa = 3.22STDD316 pKa = 3.33NCPTIINSSQLDD328 pKa = 3.61TDD330 pKa = 3.69KK331 pKa = 11.62DD332 pKa = 4.04GLGDD336 pKa = 3.77EE337 pKa = 5.06CDD339 pKa = 4.04EE340 pKa = 6.11DD341 pKa = 5.22DD342 pKa = 5.9DD343 pKa = 5.67NDD345 pKa = 5.36GILDD349 pKa = 4.79LLPPGPDD356 pKa = 2.92NCRR359 pKa = 11.84LVPNPGQEE367 pKa = 4.18DD368 pKa = 4.25DD369 pKa = 4.42NGDD372 pKa = 3.69GVGDD376 pKa = 3.48ICEE379 pKa = 4.29SDD381 pKa = 3.45FDD383 pKa = 3.9QDD385 pKa = 3.89TVIDD389 pKa = 4.23QIDD392 pKa = 3.82VCPEE396 pKa = 3.54NAEE399 pKa = 3.78ITLTDD404 pKa = 3.83FRR406 pKa = 11.84AYY408 pKa = 8.19QTVVLDD414 pKa = 4.22PEE416 pKa = 5.32GDD418 pKa = 3.65AQIDD422 pKa = 4.13PNWVVLNQGMEE433 pKa = 4.11IVQTMNSDD441 pKa = 3.19PGLAVGYY448 pKa = 7.31TAFNGVDD455 pKa = 3.99FEE457 pKa = 4.97GTFHH461 pKa = 6.81VNTVTDD467 pKa = 3.57DD468 pKa = 4.08DD469 pKa = 4.35YY470 pKa = 12.0AGFIFGYY477 pKa = 9.58QDD479 pKa = 2.83SSSFYY484 pKa = 10.47VVMWKK489 pKa = 7.78QTEE492 pKa = 3.86QTYY495 pKa = 8.13WQATPFRR502 pKa = 11.84AVAEE506 pKa = 4.17PGIQLKK512 pKa = 10.07AVKK515 pKa = 10.09SKK517 pKa = 9.55TGPGEE522 pKa = 3.82HH523 pKa = 6.52LRR525 pKa = 11.84NSLWHH530 pKa = 6.51TGDD533 pKa = 3.06TSDD536 pKa = 5.19QVRR539 pKa = 11.84LLWKK543 pKa = 10.3DD544 pKa = 3.22PRR546 pKa = 11.84NVGWKK551 pKa = 10.39DD552 pKa = 3.18KK553 pKa = 11.21VSYY556 pKa = 10.33RR557 pKa = 11.84WFLQHH562 pKa = 6.9RR563 pKa = 11.84PQIGYY568 pKa = 9.07IRR570 pKa = 11.84ARR572 pKa = 11.84FYY574 pKa = 11.09EE575 pKa = 4.54GSDD578 pKa = 3.39LVADD582 pKa = 3.84SGVTIDD588 pKa = 3.09TTMRR592 pKa = 11.84GGRR595 pKa = 11.84LGVFCFSQEE604 pKa = 3.94NIIWSNLKK612 pKa = 9.68YY613 pKa = 10.31RR614 pKa = 11.84CNDD617 pKa = 4.18TIPEE621 pKa = 4.31DD622 pKa = 3.77FQEE625 pKa = 4.53FQAQQFGVGDD635 pKa = 3.49II636 pKa = 4.15
Molecular weight: 69.66 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A091KCL2|A0A091KCL2_9GRUI Stanniocalcin-1 (Fragment) OS=Chlamydotis macqueenii OX=187382 GN=N324_02285 PE=3 SV=1
DDD2 pKa = 3.69EE3 pKa = 5.64DD4 pKa = 4.23LVRR7 pKa = 11.84GLSVLCNLANQLYYY21 pKa = 8.69YY22 pKa = 10.2CEEE25 pKa = 4.04HH26 pKa = 6.39AWAADDD32 pKa = 3.38GIIRR36 pKa = 11.84VSSQKKK42 pKa = 8.66WTLSTALWAFSLLLGLLRR60 pKa = 11.84SLRR63 pKa = 11.84ILFRR67 pKa = 11.84LRR69 pKa = 11.84RR70 pKa = 11.84KKK72 pKa = 9.14QQHHH76 pKa = 5.76KK77 pKa = 9.88ASPPLSRR84 pKa = 11.84KKK86 pKa = 5.44EE87 pKa = 3.9KKK89 pKa = 10.51RR90 pKa = 11.84VQAEEE95 pKa = 4.26LSILTDDD102 pKa = 3.62ADDD105 pKa = 3.9ANAIHHH111 pKa = 7.0LPAGFLWAGRR121 pKa = 11.84LPAWSVGLLGTISSLIGIYYY141 pKa = 9.57AARR144 pKa = 11.84GRR146 pKa = 11.84NS
DDD2 pKa = 3.69EE3 pKa = 5.64DD4 pKa = 4.23LVRR7 pKa = 11.84GLSVLCNLANQLYYY21 pKa = 8.69YY22 pKa = 10.2CEEE25 pKa = 4.04HH26 pKa = 6.39AWAADDD32 pKa = 3.38GIIRR36 pKa = 11.84VSSQKKK42 pKa = 8.66WTLSTALWAFSLLLGLLRR60 pKa = 11.84SLRR63 pKa = 11.84ILFRR67 pKa = 11.84LRR69 pKa = 11.84RR70 pKa = 11.84KKK72 pKa = 9.14QQHHH76 pKa = 5.76KK77 pKa = 9.88ASPPLSRR84 pKa = 11.84KKK86 pKa = 5.44EE87 pKa = 3.9KKK89 pKa = 10.51RR90 pKa = 11.84VQAEEE95 pKa = 4.26LSILTDDD102 pKa = 3.62ADDD105 pKa = 3.9ANAIHHH111 pKa = 7.0LPAGFLWAGRR121 pKa = 11.84LPAWSVGLLGTISSLIGIYYY141 pKa = 9.57AARR144 pKa = 11.84GRR146 pKa = 11.84NS
Molecular weight: 16.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3590270 |
33 |
5242 |
433.8 |
48.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.38 ± 0.022 | 2.232 ± 0.023 |
5.043 ± 0.018 | 7.232 ± 0.032 |
3.942 ± 0.02 | 5.767 ± 0.035 |
2.523 ± 0.014 | 5.046 ± 0.019 |
6.495 ± 0.029 | 9.739 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.256 ± 0.011 | 4.185 ± 0.017 |
5.219 ± 0.034 | 4.692 ± 0.025 |
5.127 ± 0.019 | 8.228 ± 0.036 |
5.407 ± 0.021 | 6.293 ± 0.021 |
1.183 ± 0.01 | 3.006 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
