
Sunshine Coast virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Sunviridae; Sunshinevirus; Reptile sunshinevirus 1
Average proteome isoelectric point is 7.06
Get precalculated fractions of proteins
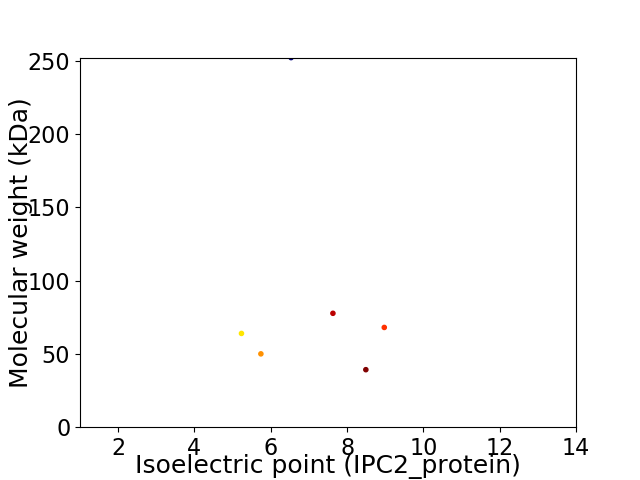
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I3VIZ0|I3VIZ0_9MONO Matrix OS=Sunshine Coast virus OX=1195087 PE=4 SV=1
MM1 pKa = 7.41EE2 pKa = 5.31LAGSVLDD9 pKa = 4.5VKK11 pKa = 9.95TQTLMEE17 pKa = 5.29GILAATKK24 pKa = 9.93EE25 pKa = 4.24DD26 pKa = 3.92TVISDD31 pKa = 3.6WSSDD35 pKa = 2.37IDD37 pKa = 3.44TAMKK41 pKa = 10.65NNSDD45 pKa = 3.66PQIPAWASGVNYY57 pKa = 10.56NIDD60 pKa = 3.39EE61 pKa = 4.77AEE63 pKa = 4.35DD64 pKa = 3.37EE65 pKa = 4.68KK66 pKa = 11.36QALDD70 pKa = 3.76CKK72 pKa = 10.51DD73 pKa = 3.28ISVEE77 pKa = 4.01EE78 pKa = 4.36YY79 pKa = 10.98LRR81 pKa = 11.84MPSAPEE87 pKa = 3.47AHH89 pKa = 7.55DD90 pKa = 3.5IPEE93 pKa = 4.31DD94 pKa = 3.64TNRR97 pKa = 11.84FSTIYY102 pKa = 10.38EE103 pKa = 3.78VDD105 pKa = 3.67EE106 pKa = 4.55EE107 pKa = 4.46CGEE110 pKa = 4.17RR111 pKa = 11.84AILSLNGEE119 pKa = 4.5DD120 pKa = 3.65TNEE123 pKa = 4.03PTGHH127 pKa = 6.71IEE129 pKa = 4.27GEE131 pKa = 4.44SGTATHH137 pKa = 6.04QGEE140 pKa = 4.28YY141 pKa = 10.21TIDD144 pKa = 3.29LGQINRR150 pKa = 11.84DD151 pKa = 3.11IKK153 pKa = 11.27NSIQTFIEE161 pKa = 4.03TGKK164 pKa = 9.81RR165 pKa = 11.84HH166 pKa = 6.48PDD168 pKa = 2.71ILAINPKK175 pKa = 10.59LFMQNAAIIWGNSAVRR191 pKa = 11.84GRR193 pKa = 11.84ANISPISFIHH203 pKa = 5.67LRR205 pKa = 11.84SGISHH210 pKa = 7.86PIKK213 pKa = 10.22SWFFLGMMMASFNKK227 pKa = 9.66KK228 pKa = 9.59GKK230 pKa = 6.88PTGRR234 pKa = 11.84AFFVPHH240 pKa = 5.4EE241 pKa = 4.25QIMAISQKK249 pKa = 11.18DD250 pKa = 3.37MQNFNKK256 pKa = 9.75VKK258 pKa = 10.53HH259 pKa = 6.41ILDD262 pKa = 3.73KK263 pKa = 11.22YY264 pKa = 11.13KK265 pKa = 10.89EE266 pKa = 4.15DD267 pKa = 3.62TRR269 pKa = 11.84QSNIFIVDD277 pKa = 3.41ITRR280 pKa = 11.84EE281 pKa = 3.69IVLNEE286 pKa = 3.59KK287 pKa = 9.29MKK289 pKa = 10.33QFGVIRR295 pKa = 11.84SYY297 pKa = 11.4ALCVPKK303 pKa = 10.45YY304 pKa = 9.57HH305 pKa = 7.01HH306 pKa = 7.2LFPSSSLNTIFGQVQSGNIRR326 pKa = 11.84MHH328 pKa = 6.6IGEE331 pKa = 4.51RR332 pKa = 11.84EE333 pKa = 4.09VIEE336 pKa = 4.82DD337 pKa = 4.12LNAKK341 pKa = 9.68CIEE344 pKa = 4.29YY345 pKa = 10.54NLSDD349 pKa = 4.66LPSDD353 pKa = 3.68VLNGEE358 pKa = 4.14RR359 pKa = 11.84EE360 pKa = 3.88EE361 pKa = 4.8DD362 pKa = 3.11KK363 pKa = 11.16SNRR366 pKa = 11.84YY367 pKa = 8.5IQDD370 pKa = 4.13DD371 pKa = 3.44MDD373 pKa = 3.85EE374 pKa = 4.24DD375 pKa = 4.08KK376 pKa = 11.02KK377 pKa = 11.22RR378 pKa = 11.84KK379 pKa = 7.78TNPTPITLNEE389 pKa = 4.01LRR391 pKa = 11.84RR392 pKa = 11.84EE393 pKa = 3.92LDD395 pKa = 3.88SISQNQTQFQEE406 pKa = 4.29IILLILKK413 pKa = 9.55KK414 pKa = 10.21QEE416 pKa = 3.64EE417 pKa = 4.74RR418 pKa = 11.84IDD420 pKa = 4.14KK421 pKa = 10.37ISEE424 pKa = 3.76ILGDD428 pKa = 3.92VLSLQQKK435 pKa = 5.45TTSQGEE441 pKa = 3.88IWLGASIKK449 pKa = 10.3AAEE452 pKa = 5.11KK453 pKa = 10.02ISQFEE458 pKa = 4.58GNQEE462 pKa = 4.05TYY464 pKa = 7.59QTYY467 pKa = 11.01LLTKK471 pKa = 10.28IMIHH475 pKa = 5.81LRR477 pKa = 11.84EE478 pKa = 4.11SEE480 pKa = 3.79QRR482 pKa = 11.84IIKK485 pKa = 7.72EE486 pKa = 3.77LCHH489 pKa = 6.46FKK491 pKa = 10.49TNPVRR496 pKa = 11.84PEE498 pKa = 3.57IPTPNLLLRR507 pKa = 11.84RR508 pKa = 11.84ALEE511 pKa = 4.12TMTEE515 pKa = 4.04EE516 pKa = 4.21MSLHH520 pKa = 6.05PKK522 pKa = 10.06EE523 pKa = 5.96NLIKK527 pKa = 9.44WNPKK531 pKa = 6.23ITEE534 pKa = 4.05IVEE537 pKa = 4.36SKK539 pKa = 10.85DD540 pKa = 3.14PEE542 pKa = 4.81RR543 pKa = 11.84INKK546 pKa = 7.3MVRR549 pKa = 11.84FMIQEE554 pKa = 4.0RR555 pKa = 11.84NKK557 pKa = 10.27YY558 pKa = 9.13
MM1 pKa = 7.41EE2 pKa = 5.31LAGSVLDD9 pKa = 4.5VKK11 pKa = 9.95TQTLMEE17 pKa = 5.29GILAATKK24 pKa = 9.93EE25 pKa = 4.24DD26 pKa = 3.92TVISDD31 pKa = 3.6WSSDD35 pKa = 2.37IDD37 pKa = 3.44TAMKK41 pKa = 10.65NNSDD45 pKa = 3.66PQIPAWASGVNYY57 pKa = 10.56NIDD60 pKa = 3.39EE61 pKa = 4.77AEE63 pKa = 4.35DD64 pKa = 3.37EE65 pKa = 4.68KK66 pKa = 11.36QALDD70 pKa = 3.76CKK72 pKa = 10.51DD73 pKa = 3.28ISVEE77 pKa = 4.01EE78 pKa = 4.36YY79 pKa = 10.98LRR81 pKa = 11.84MPSAPEE87 pKa = 3.47AHH89 pKa = 7.55DD90 pKa = 3.5IPEE93 pKa = 4.31DD94 pKa = 3.64TNRR97 pKa = 11.84FSTIYY102 pKa = 10.38EE103 pKa = 3.78VDD105 pKa = 3.67EE106 pKa = 4.55EE107 pKa = 4.46CGEE110 pKa = 4.17RR111 pKa = 11.84AILSLNGEE119 pKa = 4.5DD120 pKa = 3.65TNEE123 pKa = 4.03PTGHH127 pKa = 6.71IEE129 pKa = 4.27GEE131 pKa = 4.44SGTATHH137 pKa = 6.04QGEE140 pKa = 4.28YY141 pKa = 10.21TIDD144 pKa = 3.29LGQINRR150 pKa = 11.84DD151 pKa = 3.11IKK153 pKa = 11.27NSIQTFIEE161 pKa = 4.03TGKK164 pKa = 9.81RR165 pKa = 11.84HH166 pKa = 6.48PDD168 pKa = 2.71ILAINPKK175 pKa = 10.59LFMQNAAIIWGNSAVRR191 pKa = 11.84GRR193 pKa = 11.84ANISPISFIHH203 pKa = 5.67LRR205 pKa = 11.84SGISHH210 pKa = 7.86PIKK213 pKa = 10.22SWFFLGMMMASFNKK227 pKa = 9.66KK228 pKa = 9.59GKK230 pKa = 6.88PTGRR234 pKa = 11.84AFFVPHH240 pKa = 5.4EE241 pKa = 4.25QIMAISQKK249 pKa = 11.18DD250 pKa = 3.37MQNFNKK256 pKa = 9.75VKK258 pKa = 10.53HH259 pKa = 6.41ILDD262 pKa = 3.73KK263 pKa = 11.22YY264 pKa = 11.13KK265 pKa = 10.89EE266 pKa = 4.15DD267 pKa = 3.62TRR269 pKa = 11.84QSNIFIVDD277 pKa = 3.41ITRR280 pKa = 11.84EE281 pKa = 3.69IVLNEE286 pKa = 3.59KK287 pKa = 9.29MKK289 pKa = 10.33QFGVIRR295 pKa = 11.84SYY297 pKa = 11.4ALCVPKK303 pKa = 10.45YY304 pKa = 9.57HH305 pKa = 7.01HH306 pKa = 7.2LFPSSSLNTIFGQVQSGNIRR326 pKa = 11.84MHH328 pKa = 6.6IGEE331 pKa = 4.51RR332 pKa = 11.84EE333 pKa = 4.09VIEE336 pKa = 4.82DD337 pKa = 4.12LNAKK341 pKa = 9.68CIEE344 pKa = 4.29YY345 pKa = 10.54NLSDD349 pKa = 4.66LPSDD353 pKa = 3.68VLNGEE358 pKa = 4.14RR359 pKa = 11.84EE360 pKa = 3.88EE361 pKa = 4.8DD362 pKa = 3.11KK363 pKa = 11.16SNRR366 pKa = 11.84YY367 pKa = 8.5IQDD370 pKa = 4.13DD371 pKa = 3.44MDD373 pKa = 3.85EE374 pKa = 4.24DD375 pKa = 4.08KK376 pKa = 11.02KK377 pKa = 11.22RR378 pKa = 11.84KK379 pKa = 7.78TNPTPITLNEE389 pKa = 4.01LRR391 pKa = 11.84RR392 pKa = 11.84EE393 pKa = 3.92LDD395 pKa = 3.88SISQNQTQFQEE406 pKa = 4.29IILLILKK413 pKa = 9.55KK414 pKa = 10.21QEE416 pKa = 3.64EE417 pKa = 4.74RR418 pKa = 11.84IDD420 pKa = 4.14KK421 pKa = 10.37ISEE424 pKa = 3.76ILGDD428 pKa = 3.92VLSLQQKK435 pKa = 5.45TTSQGEE441 pKa = 3.88IWLGASIKK449 pKa = 10.3AAEE452 pKa = 5.11KK453 pKa = 10.02ISQFEE458 pKa = 4.58GNQEE462 pKa = 4.05TYY464 pKa = 7.59QTYY467 pKa = 11.01LLTKK471 pKa = 10.28IMIHH475 pKa = 5.81LRR477 pKa = 11.84EE478 pKa = 4.11SEE480 pKa = 3.79QRR482 pKa = 11.84IIKK485 pKa = 7.72EE486 pKa = 3.77LCHH489 pKa = 6.46FKK491 pKa = 10.49TNPVRR496 pKa = 11.84PEE498 pKa = 3.57IPTPNLLLRR507 pKa = 11.84RR508 pKa = 11.84ALEE511 pKa = 4.12TMTEE515 pKa = 4.04EE516 pKa = 4.21MSLHH520 pKa = 6.05PKK522 pKa = 10.06EE523 pKa = 5.96NLIKK527 pKa = 9.44WNPKK531 pKa = 6.23ITEE534 pKa = 4.05IVEE537 pKa = 4.36SKK539 pKa = 10.85DD540 pKa = 3.14PEE542 pKa = 4.81RR543 pKa = 11.84INKK546 pKa = 7.3MVRR549 pKa = 11.84FMIQEE554 pKa = 4.0RR555 pKa = 11.84NKK557 pKa = 10.27YY558 pKa = 9.13
Molecular weight: 63.98 kDa
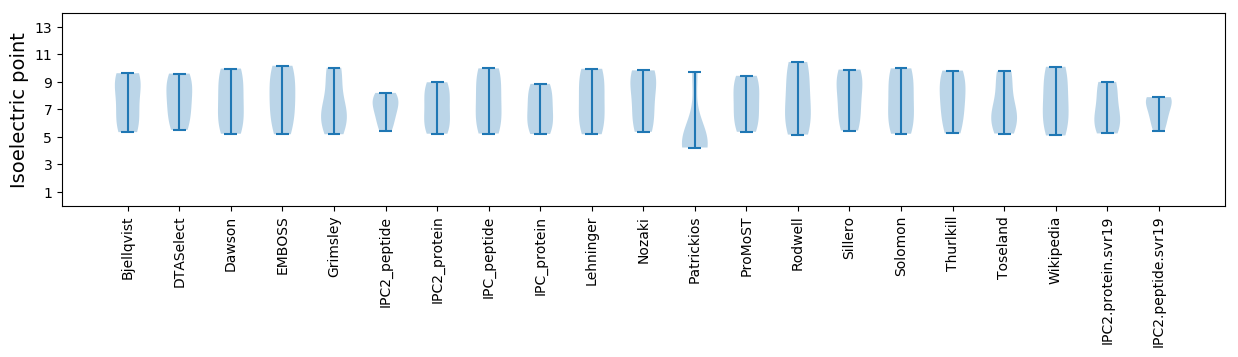
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I3VIZ2|I3VIZ2_9MONO Putative attachment protein OS=Sunshine Coast virus OX=1195087 PE=4 SV=1
MM1 pKa = 7.66EE2 pKa = 4.7NQHH5 pKa = 7.62IIIKK9 pKa = 9.97KK10 pKa = 8.41KK11 pKa = 10.61KK12 pKa = 9.27KK13 pKa = 9.58GKK15 pKa = 9.65KK16 pKa = 8.37GKK18 pKa = 9.76KK19 pKa = 8.55NKK21 pKa = 9.79KK22 pKa = 7.18EE23 pKa = 3.82RR24 pKa = 11.84KK25 pKa = 9.23KK26 pKa = 10.2KK27 pKa = 9.94GKK29 pKa = 9.76RR30 pKa = 11.84EE31 pKa = 3.74KK32 pKa = 10.48EE33 pKa = 3.97RR34 pKa = 11.84EE35 pKa = 4.17KK36 pKa = 11.02KK37 pKa = 9.02STPKK41 pKa = 10.14PQLKK45 pKa = 10.13PKK47 pKa = 10.24ILTNLRR53 pKa = 11.84GDD55 pKa = 4.04SPMEE59 pKa = 4.01TPQLFSGRR67 pKa = 11.84TGHH70 pKa = 5.1QWSRR74 pKa = 11.84RR75 pKa = 11.84YY76 pKa = 9.58IRR78 pKa = 11.84YY79 pKa = 10.06NYY81 pKa = 10.2FLTLTLLTVIFADD94 pKa = 4.03HH95 pKa = 6.98QEE97 pKa = 4.05LTQNSDD103 pKa = 3.59IIVSTVALEE112 pKa = 4.07TRR114 pKa = 11.84VVSVQTGSSWFLLRR128 pKa = 11.84VNTTFKK134 pKa = 11.21NDD136 pKa = 3.33GLTSRR141 pKa = 11.84CTKK144 pKa = 10.35LGEE147 pKa = 4.18NFVSHH152 pKa = 6.24VNEE155 pKa = 3.8FQEE158 pKa = 4.44NIKK161 pKa = 8.63ITASSLIPKK170 pKa = 9.52QAILEE175 pKa = 4.34VNTAKK180 pKa = 10.92NLVSALTRR188 pKa = 11.84TTRR191 pKa = 11.84SNRR194 pKa = 11.84RR195 pKa = 11.84KK196 pKa = 9.61RR197 pKa = 11.84LAGAAIAVAAVAISGAALVTAGIALAEE224 pKa = 4.21TRR226 pKa = 11.84TLAASVDD233 pKa = 4.19SVISGAEE240 pKa = 3.81KK241 pKa = 10.29TNAALNQLTQTMKK254 pKa = 10.89SFTKK258 pKa = 9.03EE259 pKa = 3.93TQDD262 pKa = 5.05NISTLAKK269 pKa = 10.03EE270 pKa = 3.66ISEE273 pKa = 4.08ANKK276 pKa = 10.55RR277 pKa = 11.84IDD279 pKa = 3.28CLEE282 pKa = 4.2YY283 pKa = 8.85YY284 pKa = 10.21TEE286 pKa = 4.27VKK288 pKa = 10.68NQFLDD293 pKa = 3.98FTSYY297 pKa = 11.05NYY299 pKa = 10.31EE300 pKa = 3.71ILQASMSKK308 pKa = 10.58NGGIPLRR315 pKa = 11.84LMKK318 pKa = 10.34RR319 pKa = 11.84IMVPNPQMTAQSNTIEE335 pKa = 4.19EE336 pKa = 4.37LVLRR340 pKa = 11.84RR341 pKa = 11.84GRR343 pKa = 11.84IRR345 pKa = 11.84LLAYY349 pKa = 8.73TNSTLTYY356 pKa = 9.54RR357 pKa = 11.84IDD359 pKa = 3.56VPRR362 pKa = 11.84FASMGKK368 pKa = 9.68ISPLLISVDD377 pKa = 3.87EE378 pKa = 4.91PISLIGSGIYY388 pKa = 9.99TSILSVDD395 pKa = 4.05LGIVLLDD402 pKa = 3.89CEE404 pKa = 4.72EE405 pKa = 4.94DD406 pKa = 3.69NISYY410 pKa = 10.06LCSSSSPSDD419 pKa = 4.28AIVKK423 pKa = 10.1NCLKK427 pKa = 10.76DD428 pKa = 3.47STADD432 pKa = 3.89CISALQEE439 pKa = 3.92SGAKK443 pKa = 8.49KK444 pKa = 10.2GKK446 pKa = 9.7YY447 pKa = 8.9FVYY450 pKa = 10.54QSGFLVSRR458 pKa = 11.84CSQQVCTCLEE468 pKa = 3.87PEE470 pKa = 3.53IRR472 pKa = 11.84RR473 pKa = 11.84IYY475 pKa = 10.24VSHH478 pKa = 7.42PLSKK482 pKa = 9.79EE483 pKa = 3.98TSQCTQVSIKK493 pKa = 9.46TQDD496 pKa = 3.01QTFRR500 pKa = 11.84LLLTSVKK507 pKa = 10.33AAGIINIQAPTEE519 pKa = 4.2IKK521 pKa = 10.32RR522 pKa = 11.84KK523 pKa = 4.99TWKK526 pKa = 10.64DD527 pKa = 2.75IGLTFGKK534 pKa = 10.56YY535 pKa = 5.87NTKK538 pKa = 10.18FEE540 pKa = 4.72EE541 pKa = 4.14IEE543 pKa = 3.95EE544 pKa = 4.74LIEE547 pKa = 4.18NSNGDD552 pKa = 3.68LEE554 pKa = 4.29KK555 pKa = 11.06ARR557 pKa = 11.84TYY559 pKa = 11.13LVLSQNLKK567 pKa = 9.37VVVIVTSIVIAFLILFVIFILKK589 pKa = 10.31RR590 pKa = 11.84IYY592 pKa = 10.12DD593 pKa = 3.91LKK595 pKa = 11.29NRR597 pKa = 11.84VTKK600 pKa = 10.47IEE602 pKa = 4.18VYY604 pKa = 10.46LEE606 pKa = 4.0RR607 pKa = 6.05
MM1 pKa = 7.66EE2 pKa = 4.7NQHH5 pKa = 7.62IIIKK9 pKa = 9.97KK10 pKa = 8.41KK11 pKa = 10.61KK12 pKa = 9.27KK13 pKa = 9.58GKK15 pKa = 9.65KK16 pKa = 8.37GKK18 pKa = 9.76KK19 pKa = 8.55NKK21 pKa = 9.79KK22 pKa = 7.18EE23 pKa = 3.82RR24 pKa = 11.84KK25 pKa = 9.23KK26 pKa = 10.2KK27 pKa = 9.94GKK29 pKa = 9.76RR30 pKa = 11.84EE31 pKa = 3.74KK32 pKa = 10.48EE33 pKa = 3.97RR34 pKa = 11.84EE35 pKa = 4.17KK36 pKa = 11.02KK37 pKa = 9.02STPKK41 pKa = 10.14PQLKK45 pKa = 10.13PKK47 pKa = 10.24ILTNLRR53 pKa = 11.84GDD55 pKa = 4.04SPMEE59 pKa = 4.01TPQLFSGRR67 pKa = 11.84TGHH70 pKa = 5.1QWSRR74 pKa = 11.84RR75 pKa = 11.84YY76 pKa = 9.58IRR78 pKa = 11.84YY79 pKa = 10.06NYY81 pKa = 10.2FLTLTLLTVIFADD94 pKa = 4.03HH95 pKa = 6.98QEE97 pKa = 4.05LTQNSDD103 pKa = 3.59IIVSTVALEE112 pKa = 4.07TRR114 pKa = 11.84VVSVQTGSSWFLLRR128 pKa = 11.84VNTTFKK134 pKa = 11.21NDD136 pKa = 3.33GLTSRR141 pKa = 11.84CTKK144 pKa = 10.35LGEE147 pKa = 4.18NFVSHH152 pKa = 6.24VNEE155 pKa = 3.8FQEE158 pKa = 4.44NIKK161 pKa = 8.63ITASSLIPKK170 pKa = 9.52QAILEE175 pKa = 4.34VNTAKK180 pKa = 10.92NLVSALTRR188 pKa = 11.84TTRR191 pKa = 11.84SNRR194 pKa = 11.84RR195 pKa = 11.84KK196 pKa = 9.61RR197 pKa = 11.84LAGAAIAVAAVAISGAALVTAGIALAEE224 pKa = 4.21TRR226 pKa = 11.84TLAASVDD233 pKa = 4.19SVISGAEE240 pKa = 3.81KK241 pKa = 10.29TNAALNQLTQTMKK254 pKa = 10.89SFTKK258 pKa = 9.03EE259 pKa = 3.93TQDD262 pKa = 5.05NISTLAKK269 pKa = 10.03EE270 pKa = 3.66ISEE273 pKa = 4.08ANKK276 pKa = 10.55RR277 pKa = 11.84IDD279 pKa = 3.28CLEE282 pKa = 4.2YY283 pKa = 8.85YY284 pKa = 10.21TEE286 pKa = 4.27VKK288 pKa = 10.68NQFLDD293 pKa = 3.98FTSYY297 pKa = 11.05NYY299 pKa = 10.31EE300 pKa = 3.71ILQASMSKK308 pKa = 10.58NGGIPLRR315 pKa = 11.84LMKK318 pKa = 10.34RR319 pKa = 11.84IMVPNPQMTAQSNTIEE335 pKa = 4.19EE336 pKa = 4.37LVLRR340 pKa = 11.84RR341 pKa = 11.84GRR343 pKa = 11.84IRR345 pKa = 11.84LLAYY349 pKa = 8.73TNSTLTYY356 pKa = 9.54RR357 pKa = 11.84IDD359 pKa = 3.56VPRR362 pKa = 11.84FASMGKK368 pKa = 9.68ISPLLISVDD377 pKa = 3.87EE378 pKa = 4.91PISLIGSGIYY388 pKa = 9.99TSILSVDD395 pKa = 4.05LGIVLLDD402 pKa = 3.89CEE404 pKa = 4.72EE405 pKa = 4.94DD406 pKa = 3.69NISYY410 pKa = 10.06LCSSSSPSDD419 pKa = 4.28AIVKK423 pKa = 10.1NCLKK427 pKa = 10.76DD428 pKa = 3.47STADD432 pKa = 3.89CISALQEE439 pKa = 3.92SGAKK443 pKa = 8.49KK444 pKa = 10.2GKK446 pKa = 9.7YY447 pKa = 8.9FVYY450 pKa = 10.54QSGFLVSRR458 pKa = 11.84CSQQVCTCLEE468 pKa = 3.87PEE470 pKa = 3.53IRR472 pKa = 11.84RR473 pKa = 11.84IYY475 pKa = 10.24VSHH478 pKa = 7.42PLSKK482 pKa = 9.79EE483 pKa = 3.98TSQCTQVSIKK493 pKa = 9.46TQDD496 pKa = 3.01QTFRR500 pKa = 11.84LLLTSVKK507 pKa = 10.33AAGIINIQAPTEE519 pKa = 4.2IKK521 pKa = 10.32RR522 pKa = 11.84KK523 pKa = 4.99TWKK526 pKa = 10.64DD527 pKa = 2.75IGLTFGKK534 pKa = 10.56YY535 pKa = 5.87NTKK538 pKa = 10.18FEE540 pKa = 4.72EE541 pKa = 4.14IEE543 pKa = 3.95EE544 pKa = 4.74LIEE547 pKa = 4.18NSNGDD552 pKa = 3.68LEE554 pKa = 4.29KK555 pKa = 11.06ARR557 pKa = 11.84TYY559 pKa = 11.13LVLSQNLKK567 pKa = 9.37VVVIVTSIVIAFLILFVIFILKK589 pKa = 10.31RR590 pKa = 11.84IYY592 pKa = 10.12DD593 pKa = 3.91LKK595 pKa = 11.29NRR597 pKa = 11.84VTKK600 pKa = 10.47IEE602 pKa = 4.18VYY604 pKa = 10.46LEE606 pKa = 4.0RR607 pKa = 6.05
Molecular weight: 68.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4847 |
361 |
2198 |
807.8 |
91.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.734 ± 0.998 | 1.754 ± 0.288 |
5.178 ± 0.389 | 6.932 ± 0.481 |
3.899 ± 0.272 | 5.178 ± 0.603 |
1.795 ± 0.282 | 9.263 ± 0.422 |
7.18 ± 0.49 | 10.543 ± 0.64 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.496 ± 0.241 | 5.178 ± 0.348 |
4.312 ± 0.523 | 3.59 ± 0.333 |
5.426 ± 0.224 | 8.686 ± 0.358 |
5.756 ± 0.486 | 5.137 ± 0.287 |
0.825 ± 0.086 | 3.136 ± 0.299 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
